Abstract
The V protein of simian virus 5 (SV5) blocks interferon signaling by targeting STAT1 for proteasome-mediated degradation. Here we present three main pieces of evidence which demonstrate that the p127 subunit (DDB1) of the UV damage-specific DNA binding protein (DDB) plays a central role in this degradation process. First, the V protein of an SV5 mutant which fails to target STAT1 for degradation does not bind DDB1. Second, mutations in the N and C termini of V which abolish the binding of V to DDB1 also prevent V from blocking interferon (IFN) signaling. Third, treatment of HeLa/SV5-V cells, which constitutively express the V protein of SV5 and thus lack STAT1, with short interfering RNAs specific for DDB1 resulted in a reduction in DDB1 levels with a concomitant increase in STAT1 levels and a restoration of IFN signaling. Furthermore, STAT1 is degraded in GM02415 (2RO) cells, which have a mutation in DDB2 (the p48 subunit of DDB) which abolishes its ability to interact with DDB1, thereby demonstrating that the role of DDB1 in STAT1 degradation is independent of its association with DDB2. Evidence is also presented which demonstrates that STAT2 is required for the degradation of STAT1 by SV5. These results suggest that DDB1, STAT1, STAT2, and V may form part of a large multiprotein complex which leads to the targeted degradation of STAT1 by the proteasome.
The importance of interferons (IFNs) in controlling virus infections can be judged by the fact that viruses unable to at least partially circumvent the IFN response are attenuated in vivo, and transgenic mice unable to respond to IFN are highly susceptible to virus infections. There are two main subtypes of IFN: alpha/beta IFN (IFN-α/β), produced as a direct consequence of virus infection, and gamma IFN (IFN-γ), which is produced by subsets of activated T lymphocytes and NK cells. IFN-α/β and IFN-γ mediate their actions by inducing the expression of overlapping sets of IFN-stimulated genes, many of which possess direct or indirect antiviral activities. IFN-α/β and IFN-γ bind to independent cell-surface receptors and activate distinct but related signal transduction pathways. Following the binding of IFN-α/β to their cognate receptor, the inactive cytoplasmic transcription factors STAT1 and STAT2 become phosphorylated on tyrosine Y701 and Y690, respectively, by receptor-associated tyrosine kinases. Phosphorylated STAT1 and STAT2 form heterodimers and migrate to the nucleus, where they become associated with p48 to form the ISGF3 complex, which activates the transcription of IFN-α/β-responsive genes. Following binding of IFN-γ to its receptor, the activated receptor-associated kinases phosphorylate STAT1 on Y701, which subsequently homodimerizes to form the GAF complex, which activates transcription of IFN-γ-responsive genes (for reviews on viruses and interferons, see references 12, 25, and 47)
Many paramyxoviruses at least partially circumvent the IFN response by blocking IFN signaling, although they achieve this by distinct molecular mechanisms (6, 7, 13, 17, 23, 24, 34, 48, 53). Thus, the V proteins encoded by simian virus 5 (SV5), mumps virus, and SV41 block IFN signaling by targeting STAT1 for degradation (7, 24, 32), while the V protein of human parainfluenza virus type 2 (hPIV2) targets STAT2 for degradation (1, 32, 34, 53). Sendai virus and hPIV3 also block IFN signaling, but in the case of Sendai virus it has been shown that this is a property of the C proteins, and the process does not necessarily require STAT degradation but rather a direct interaction between C and STAT1 (6, 9, 10, 13, 17, 22, 40, 53). Evidence that the degradation of STAT1 by SV5 is mediated via the proteasome was provided by demonstrating that the proteasome inhibitors MG132 and lactacystein blocked the degradation process. All cellular components required for degradation are constitutively present within cells, since SV5-mediated degradation does not require either de novo cellular transcription or protein synthesis (7). In addition, STAT1 degradation is independent of IFN signaling, since SV5 induces the degradation of both phosphorylated and nonphosphorylated forms of STAT1 (1). Furthermore, STAT1 and STAT2 are degraded by SV5 and hPIV2, respectively, in cells deficient in the IFN signaling pathway (35). It appears that both the N-terminal P/V common domain and the cysteine-rich C-terminal domain of V are essential in the degradation process. Evidence for a role for the N-terminal domain comes from the observation that a single amino acid substitution at amino acid 100 influences the ability of the V protein of SV5 to target STAT1 for degradation in cells from different species (52). Furthermore, the differing abilities of two canine isolates of SV5, termed CPI+ (which targets STAT1 for degradation) and CPI− (which fails to degrade STAT1), have been mapped to three amino acid differences in the N-terminal P/V common domain (3). Evidence that the C terminus is also required in circumventing the IFN response comes from observations that expression of the C-terminal cysteine-rich region of the V protein of mumps virus inhibits the induction of an IFN-induced antiviral state (24) and that a recombinant hPIV2 virus that encodes a C-terminally truncated version of the V protein was sensitive to inhibition by IFN (18).
As with many other virus proteins, it appears that the V protein of SV5 may be a multifunctional protein. As well as targeting STAT1 for degradation, V binds to soluble nucleocapsid protein (NP) but not to polymeric NP and may thus have a role in virus transcription, replication, or encapsidation (39). However, its role in targeting STAT1 for degradation is not dependent upon its interaction with NP. Indeed, cells (termed 2f/SV5-V cells) which constitutively express the V protein of SV5 in the absence of other virus proteins lack STAT1 and do not respond to either IFN-α/β or IFN-γ (1). Similarly, cells which express the V protein of hPIV2 (termed 2f/PIV-V cells) lack STAT2 and do not respond to IFN-α/β, although they can respond to IFN-γ (1). It has also been shown that the V proteins of many paramyxoviruses, including SV5 and hPIV2, bind to the 127-kDa subunit (DDB1) of the UV damage DNA binding protein (27), and a role for V in slowing the cell cycle to the possible advantage of virus replication has been proposed (26).
Here we provide evidence that the interaction of V with DDB1 is essential for the degradation of STAT1 by SV5, and we demonstrate that the presence of both STAT1 and STAT2 is required for the degradation of STAT1 or STAT2 by SV5 or hPIV2, respectively.
MATERIALS AND METHODS
Cells, viruses, and IFN.
Human 2fTGH cells (30, 37), HeLa cells and their derivatives, 293 cells, and GM02415 cells (14) were grown as monolayers in 25- or 75-cm2 tissue culture flasks in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (growth medium). All cell lines were negative for mycoplasmas as screened by 4′,6′-diamidino-2-phenylindole staining. SV5 (strain W3A) (5) and hPIV2 were grown and titrated under appropriate conditions in Vero cells using maintenance medium.
For transfections, cells at 50 to 70% confluence were transfected with appropriate DNAs (1 μg of plasmid with 1.5 μl of Fugene 6 [Roche] according to the manufacturer's instructions). To isolate cells expressing the V protein, transfected cells were cultured in the presence of 400 μg of Geneticin (G418; Sigma)/ml, and resistant colonies were isolated as previously described (1). For IFN-signaling assays, HeLa cells were transiently transfected as previously described (21). At various times posttransfection, cells were or were not induced with 1,000 IU of IFN-α (Intron A; Schering-Plough)/ml for 4 h immediately prior to harvesting. Luciferase and β-galactosidase levels were determined (20), and luciferase readings were divided by β-galactosidase readings for each sample.
Plasmids.
The plasmid directing the expression of the SV5 V protein in mammalian cells, pEF.SV5-V, has been previously described (1, 7). pEF− plasmids directing the expression of C-terminal truncations of SV5 V were obtained by cloning NcoI-PstI (amino acids 1 to 56; pEF.SV5-V[1-56]), NcoI-ClaI (amino acids 1 to 157; pEF.SV5-V[1-157]), or NcoI-ScaI (amino acids 1 to 174; pEF.SV5-V[1-174]) fragments between the NcoI site and a filled-in EcoRI site of pEFplink2 (a kind gift from R. H. Treisman, Imperial Cancer Research Fund). pEF− plasmids directing the expression of N-terminal truncations of SV5 V were obtained by cloning PCR-amplified fragments (incorporating NcoI and EcoRI sites into the 5′ and 3′ ends, respectively, of the SV5 V fragment) directly between the NcoI and EcoRI sites of pEFplink2 to generate pEF.SV5-V[20-222] (amino acids 20 to 222), pEF.SV5-V[85-222] (amino acids 85 to 222), and pEF.SV5-V[104-222] (amino acids 104 to 222). Point mutants were introduced into pEF.SV5-V using recombinant PCR to create pEF.SV5-V[C193A], pEF.SV5-V[C207A], and pEF.SV5-V[C214A]; All V mutated genes were sequenced (Lark Technologies), and a double mutant, pEF.SV5-V[C193Y/C214A], was obtained as a by-product of one of the mutagenesis experiments. The IFN-α/β- and IFN-γ-responsive luciferase reporter plasmids, p(9-27ISRE)4tkΔ(−39)lucter and p(GAS)2tkΔ(−39)lucter, and the constitutive β-galactosidase reporter plasmid, pJATlacZ, have been previously described (7, 21).
The construction of the plasmid directing the stable expression of the SV5 V protein in mammalian cells, pEF.SV5-V(W3).IRES.neo, has been previously described (1). The equivalent plasmids directing the expression of the N-terminally tagged V proteins of CPI+ and CPI− were constructed in two stages. First, to introduce a c-myc 9E10 epitope tag onto the N terminus of the V proteins, the V genes were PCR amplified from pEF/CPI+ and pEF/CPI− plasmids (3) by using appropriate primers, and the fragment were inserted into the pEFlink2 vector between the NcoI and EcoRI sites to generate pEF.SV5-V(CPI+/m) and pEF.SV5-V(CPI−/m). Second, ApaI-EcoRI fragments containing the tagged V genes were excised from these plasmids and cloned between the ApaI and SmaI sites of pEF.IRES.neo (1) to generate pEF.SV5-V(CPI+/m).IRES.neo and pEF.SV5-V(CPI-/m).IRES.neo.
The plasmid directing the expression of N-terminally c-myc 9E10 epitope-tagged STAT1α protein in mammalian cells, pEF.STAT1α/m, was constructed from pEF.STAT1α (21) by PCR-directed mutagenesis. A plasmid directing STAT2 expression in mammalian cells was obtained from George Stark (Cleveland Clinic).
The plasmid directing the expression of the SV5 V protein as a GAL4 DNA binding domain-fusion protein in yeast, pGBT9.SV5-V, was constructed by inserting an NcoI-SalI fragment containing the full-length V gene between the NcoI and SalI sites of pGBT9 (2). Plasmids expressing mutant forms of SV5 V fused to the yeast GAL4 DNA binding domain were constructed from pGBT9.SV5-V or its pGBKT7 (Clontech) equivalent by subcloning the appropriate fragment from the pEF− plasmid series described above. The plasmid directing the expression of the p127 protein as a GAL4 transcriptional activation domain fusion protein in yeast, pHON3.p127, was constructed by inserting a partial AflIII (5′ end)-BamHI fragment from pCEPp127 (a kind gift from Vesna Rapic-Otrin, University of Pittsburgh) containing the entire open reading frame of p127 between the NcoI and BamHI sites of pHON3 (derived from pGAD424 [2] by replacing the partial-length ADH promoter with the full-length ADH promoter from pAS2-1). The same p127 fragment was inserted between the NcoI and BamHI sites of pGBKT7 (Clontech) to create pGBKT7p127, a plasmid used for in vitro synthesis of recombinant p127.
Plasmids directing the synthesis of Schistosoma japonicum glutathione S-transferase fused to the V proteins of SV5 (W3), CPI+ or CPI−, were constructed from the pEF− mammalian expression plasmids (3, 7) using PCR to create V gene fragments flanked by EcoRI sites, which were then cloned in-frame directly into pGEX-2TK (Amersham Biosciences).
Yeast two-hybrid assay.
GAL4 DNA binding domain/SV5 V fusion plasmids (pGBT9 or pGBKT7) were introduced into Saccharomyces cerevisiae strain CG1945 (Mata ura3-52 his3-200 lys2-801 ade2-101 trp1-901 leu2-3 −112 gal4-542 gal80-538 cyhr2 LYS2::GAL1UAS-GAL1TATA-HIS3 URA3::GAL4(17-mer)3-CYC1TATA-lacZ) (Clontech) using lithium acetate-polyethylene glycol-mediated transformation (11), and positive transformants were selected on synthetic dropout medium (SDM) lacking tryptophan. The transformants were tested for background transactivation by growth on SDM lacking histidine. Plasmids expressing full-length SV5 V exhibited a small degree of growth under these conditions, which could be effectively suppressed by the incorporation of 5 mM 3-aminotriazole. This background growth could not be suppressed in other commonly used two-hybrid strains of yeast, such as PJ69-4a (15). The GAL4 activation domain/p127 fusion plasmid, pHON3.p127, was introduced into the CG1945 strain containing the bait plasmid by using lithium acetate-polyethylene glycol-mediated transformation, and double transformants were selected on SDM lacking tryptophan and leucine. Positive interactions were scored by growth on SDM lacking histidine and supplemented with 5 mM 3-aminotriazole.
Preparation of radiolabeled antigen extracts, immunoprecipitation, and SDS-PAGE.
Cells were metabolically labeled with l-[35S]methionine (500 Ci/mmol; Amersham International Ltd., Little Chalfont, United Kingdom) for various periods in methionine-free tissue culture medium. At the end of the labeling interval, the cells were washed in ice-cold phosphate-buffered saline and lysed into immune precipitation buffer (10 mM Tris-HCl [pH 7.8], 5 mM-EDTA, 0.5% Nonidet-P40, 0.65 M NaCl) (4 × 106 to 6 × 106 cells per ml of buffer) by sonication with an ultrasonic probe. Soluble antigen extracts were obtained after pelleting the particulate material from the total cell antigen extracts by centrifugation at 12,000 × g for 30 min. Immune complexes were formed by incubation (for 2 h at 4°C). Samples (0.2 to 1 ml) of the soluble antigen extracts with an excess of anti-STAT1 C-terminal STAT1 monoclonal antibodies (MAbs) (Transduction Laboratories), a MAb (9E10) that binds to the myc epitope, a MAb that recognizes the Pk determinant in the N-terminal P/V common domain of V (anti-SV5-Pk) (46), or a MAb (V mAb 11) that recognizes an epitope in the cysteine-rich V-unique domain of V (36). The immune complexes were isolated using an excess of protein G Sepharose 4B fast flow (Sigma) (20 μl of a 50% [wt/vol] suspension per microliter of concentrated tissue culture fluid for 1 h at 4°C). The proteins in the immune complexes were dissociated by heating (100°C for 5 min) in gel electrophoresis sample buffer (0.05 M Tris-HCl [pH 7.0], 0.2% sodium dodecyl sulfate [SDS], 5% 2-mercaptoethanol, and 5% glycerol) and analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) using a 10% gel. After electrophoresis, gels were fixed, stained, and dried; labeled polypeptides were visualized by phosphorimager analysis.
Immunoblotting.
At the time of harvest, cells were washed twice with phosphate-buffered saline, disrupted into SDS gel loading buffer, sonicated, and boiled for 5 min. Polypeptides were separated by SDS-PAGE using a 7% gel and transferred to nitrocellulose membranes; STAT1 and STAT2 were detected with either a polyclonal anti-STAT1 antibody raised against the N-terminal 194 amino acids or a MAb to the N-terminal region of STAT2 (Transduction Laboratories), the V protein was detected with either the anti-Pk MAb or V mAb 11, and myc-tagged proteins were detected with the 9E10 MAb. All protein-antibody interactions were detected by enhanced chemiluminescence using horseradish peroxidase-conjugated sheep anti-mouse or donkey anti-rabbit immunoglobulin G (Amersham International Ltd.).
siRNA silencing.
The short interfering RNA (siRNA) sequence used for silencing of the p127 subunit of UV-DDB (DDB1) corresponded to the 48-67 coding sequence numbered from the start codon. Single-stranded sense RNA was used as the negative control. RNA oligonucleotides, purchased from Dharmacon Research Inc., were desalted and deprotected. Annealing of siRNA oligonucleotides to form the siRNA duplex was carried out in annealing buffer (100 mM potassium acetate, 30 mM HEPES-KOH [pH 7.4], 2 mM magnesium acetate). Templates were mixed in equimolar amounts, heated for 1 min at 95°C, and then cooled and annealed for 1 h at 37°C. Six hundred picomoles of siRNA was used to transfect 25-cm2 flasks of HeLa or HeLa/SV5-V cells by using Oligofectamine (Invitrogen) as described by Elbashir and colleagues (8). Cells were transfected twice, with a 24-h interval between transfections, and were labeled and lysed at 120 h posttransfection.
RESULTS
The presence of both STAT1 and STAT2 is necessary for the targeted degradation of STAT1 and STAT2 by SV5 and hPIV2, respectively.
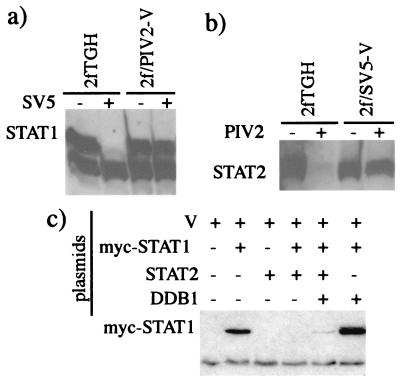
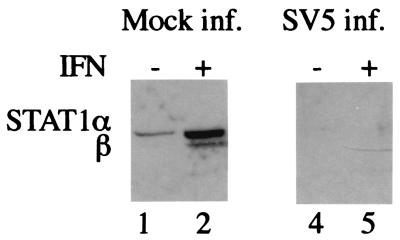
Comparing the replication of SV5 and hPIV2 in the 2fTGH cells with that in 2f/SV5-V (which lack STAT1) and 2f/PIV2-V cells (which lack STAT2) (1), we noted that SV5 induced the degradation of STAT1 in 2fTGH cells but not in 2f/PIV2-V cells (Fig. 1a). Similarly, hPIV2 induced the degradation of STAT2 in 2fTGH cells but not in 2f/SV5-V cells (Fig. 1b). One explanation for these results was that the presence of both STAT1 and STAT2 is required in the degradation process by SV5 or hPIV2, respectively. Alternatively, the endogenously expressed V proteins in 2f/SV5-V and 2f/PIV2-V cells may have sequestered a host cell protein(s) required for STAT degradation upon subsequent infection with virus. One possible candidate for such a host cell protein was DDB1, since it is known to bind strongly to V (27). To test this, 293 cells (because 2fTGH cells transfect very inefficiently) were transiently transfected with combinations of plasmids expressing the V protein of SV5, myc-tagged STAT1 (to distinguish it from endogenous STAT1), STAT2, or DDB1. It can be seen from Fig. 1c that myc-STAT1 was degraded only if V, myc-STAT1, and STAT2 were coexpressed, confirming the requirement for STAT2 in the degradation of STAT1 by the V protein of SV5. Presumably, since STAT2 is required for STAT1 turnover, the low levels of endogenous STAT2 in 293 cells were insufficient for the increased turnover required to see a loss of overexpressed STAT1 in transiently transfected cells. Overexpression of DDB1 had a small inhibitory effect on STAT1 degradation. The reason for this was unclear, but we speculated that it may have been because overexpression of DDB1 resulted in the sequestration of limiting amounts of another host cell protein(s) required for STAT1 degradation. If this was the case, although somewhat counterintuitive, it may also have meant that DDB1 had a role to play in STAT1 degradation. We therefore decided to examine in greater detail the role of DDB1 in STAT1 degradation.
FIG. 1.
Panels a and b, respectively; 2fTGH, 2f/PIV2-V, and 2f/SV5-V cells were (+) or were not (-) infected with 5 PFU of SV5/cell (a) or hPIV2/cell (b) as indicated. At 16 h p.i. the cells were harvested, and the presence of STAT1 or STAT2 was detected by immunoblot analysis. (c) 293 cells were transiently transfected with combinations of plasmids expressing V, myc-STAT1, STAT2, and DDB1 (as shown), with a total of 8 μg of plasmid per transfection, 2 μg of each plasmid together with the appropriate amount of carrier plasmid (pEFplink2). At 48 h posttransfection the cells were harvested, and the presence of myc-STAT1 was detected by immunoblot analysis.
Involvement of DDB1 in the degradation of STAT1 by SV5.
CPI+ and CPI− are two canine strains of SV5 that differ in their ability to block IFN signaling; CPI+ blocks IFN signaling by targeting STAT1 for degradation, while CPI− fails to target STAT1 for degradation (3). Although it has been reported that the V protein of the W3 strain of SV5 binds DDB1 (1, 27), it was not known whether DDB1 also binds to the V proteins of CPI+ and CPI−. To examine this, cell lines that constitutively express the V proteins of CPI+ and CPI− were derived from 2fTGH cells as previously described (3). These lines were termed 2f/SV5-V(CPI+) and 2f/SV5-V(CPI−), respectively, to distinguish them from the original cell line, now termed 2f/SV5-V(W3), which was constructed using the V gene derived from the W3 strain of SV5. In addition, since the anti-Pk MAb used to detect the V protein of the W3 and CPI+ isolates of SV5 fails to interact with the V protein of CPI− due to a single amino acid substitution in the Pk epitope in CPI− (46; see also Fig. 4a), we constructed two additional cell lines in which the myc tag was added to the N terminus of CPI+ and CPI−; these cell lines were 2f/SV5-V(CPI+/m) and 2f/SV5-V(CPI-/m), respectively.
FIG. 4.
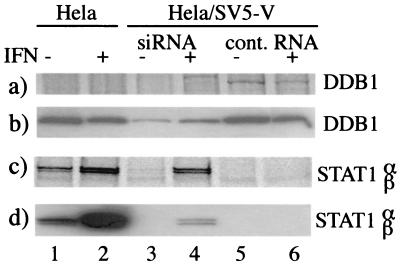
HeLa and HeLa/SV5-V cells were or were not treated with siRNA or control RNA twice at 24-h intervals. At 100 h posttreatment, IFN was or was not added to the culture medium, and 20 h later the cells were metabolically labeled with [35S]methionine for 5 h in the presence or absence of IFN as appropriate. Cells were then harvested, and the levels of DDB1 estimated by coimmunoprecipitation with V (a) (note that DDB1 was detected in HeLa/SV5-V cells treated with control RNA but not when they were treated with siRNAs and that DDB1 was not immunoprecipitated from HeLa cells as they do not express V) and immunoblot analysis (b). Levels of STAT1 in the cell samples were also examined by immunoprecipitation (c) and immunoblot analysis (d). Note the increase in levels of STAT1 in HeLa/SV5-V cells treated with siRNAs, especially following incubation with IFN (lanes 3 and 4), but not in cells treated with control RNA (lanes 5 and 6).
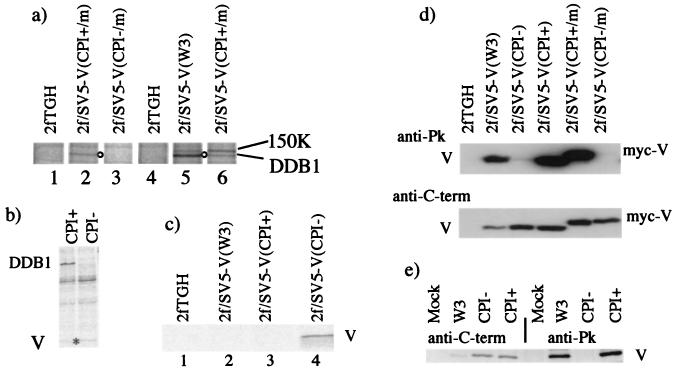
In a series of experiments to examine the coimmune precipitation of host cell proteins with the V proteins of these viruses, it became evident that DDB1 and an as yet unidentified host cell protein of 150 kDa (1) were coprecipitated with the V proteins of W3 and CPI+ (Fig. 2a, lanes 5 and 6, respectively). However, the V protein of CPI− failed to precipitate DDB1 (Fig. 2a, compare lanes 2 and 3). To confirm that the DDB1 protein was not coprecipitated with the V protein of CPI−, 293 cells were also transiently transfected with plasmids expressing myc-tagged V proteins of either CPI+ or CPI−. While clearly detectable levels of DDB1 were precipitated by the V protein of CPI+, no detectable DDB1 was coprecipitated with the V protein of CPI− (Fig. 2b). The ability of glutathione S-transferase-V fusion proteins to interact with in vitro-translated, [35S]methionine-labeled DDB1 was also examined. These experiments again demonstrated that while the V proteins of W3 and CPI+ interact with DDB1, the V protein of CPI− binds poorly, if at all, to DDB1 (data not shown).
FIG. 2.
(a) DDB1 is coimmunoprecipitated with the V protein of the W3 and CPI+ strains of SV5 but not the CPI− strain. Flasks (75 cm2) of 2fTGH, 2f/SV5-V(CPI+/m), 2fSV5-V(CPI-/m), and 2f/SV5-V(W3) cells were metabolically labeled with [35S]methionine for 5 h, harvested, and immunoprecipitated with anti-myc (lanes 1 to 3) or anti-Pk (lanes 4 to 6) antibodies. (b) DDB1 is coimmunoprecipitated with CPI+ V but not CPI− V in 293 cells transiently transfected with plasmids expressing DDB1 together with CPI+ or CPI− myc-tagged V (2 μg of each plasmid per 25-cm2 flask). Cells were labeled with [35S]methionine for 4 h at 48 h posttransfection, harvested, and immunoprecipitated with anti-myc antibody. The position of V is indicated with an asterisk. (c) The anti-C-terminus antibody (V mAb 11) immunoprecipitates the V protein from 2f/SV5-V(CPI−) cells but not from 2f/SV5(W3) or 2f/SV5(CPI+) cells. Cells were labeled with [35S]methionine for 4 h, harvested, and immunoprecipitated with V mAb 11 antibody. (d) Immunoblot assays of total cell lysates of the cells reveals that the anti-Pk MAb interacts with the V proteins of W3 and CPI+ but not CPI− while the anti-C-term antibody (V mAb 11) interacts with the V proteins of W3, CPI+, and CPI−. (e) The anti-C terminal (V mAb 11) antibody immunoprecipitates V proteins of W3, CPI+, and CPI− from infected cell lysates. 2fTGH cells were mock infected or were infected with SV5 strains W3, CPI+, or CPI− at a multiplicity of infection of 1, and at 16 h postinfection they were metabolically labeled for 4 h with [35S]methionine and immunoprecipitated with anti-C-terminus (V mAb 11) or anti-Pk MAbs as indicated.
W3, CPI+ and CPI− have identical C-terminal domains and thus react in immunoblots with an antibody (V mAb 11; anti-C terminus) that recognizes a determinant in the conserved V-unique, C-terminal domain (Fig. 2d). It was therefore surprising that while the V mAb 11 antibody immunoprecipitated the V protein from 2f/SV5-V(CPI−) cells, it failed to immunoprecipitate the V protein from 2f/SV5-V(W3) or 2f/SV5-V(CPI+) cells (Fig. 2c). The binding of V mAb 11 to the V protein of W3 and CPI+ may thus be sterically blocked in 2f/SV5-V(W3) and 2f/SV5-V(CPI+) cells by its interaction with a host cell protein, such as DDB1. If this was the case, then in virus-infected cells, in which the V protein is made in much greater amounts than in the stable cell lines, V may be in excess over the host cell protein(s), and consequently there would be a subpopulation of V which could be precipitated by the V mAb 11 antibody. To test this, the ability of the V mAb 11 antibody to immunoprecipitate V from 2fTGH cells infected with W3, CPI+, or CPI− was examined. It can be seen from Fig. 2e that the V mAb 11 antibody did indeed precipitate V from cells infected with all three viruses, confirming that the inability of V mAb 11 to precipitate V from 2f/SV5-V(W3) and 2f/SV5-V(CPI+) was not due to its intrinsic inability to recognize the V proteins of these viruses.
Mapping the interaction site for DDB1 on the SV5 V protein; correlation with a block to IFN signaling.
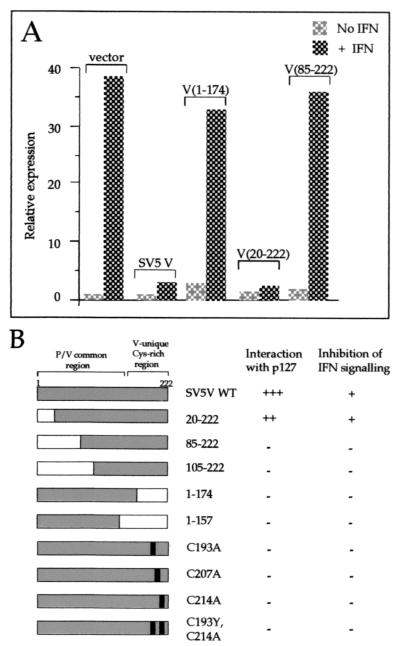
It has been previously shown that the cysteine residues in the C terminus of V are essential for its interaction with DDB1 (27). We therefore decided to determine whether these residues are also required for the SV5 V protein to block IFN signaling and whether there is tight correlation between binding DDB1 and the ability of V to block IFN signaling. To this end, a series of N- and C-terminally truncated versions of V were constructed, and a number of point mutations of conserved cysteine residues were made in the C-terminal V-unique domain. These mutants of V were tested for their ability to bind DDB1 in a yeast two-hybrid screen and for their ability to block IFN signaling (Fig. 3). From these results, it was clear that any deletion in the C terminus of V or, in agreement with the results of Lin et al. (27), mutations of the conserved cysteine residues at positions 193, 207, and 214 prevented V from binding DDB1. In addition, although the first 20 amino acids at the N terminus of V could be deleted without effecting DDB1 binding, deletion of the first 85 amino acids abolished binding. There was striking correlation between the loss of ability of truncated V to bind DDB1 and a loss in its ability to block IFN signaling (Fig. 3, panel b).
FIG. 3.
(A) Examples of the ability of altered SV5 V proteins to block IFN signaling are shown. HeLa cells were transfected with plasmids as indicated, a luciferase reporter plasmid, and a β-galactosidase expression vector. Cells were or were not treated with 1,000 U of IFN-á for 4 h; luciferase levels in extracts were normalized to β-galactosidase levels, and the relative expression values were plotted; a value of 1.0 was assigned to the basal expression level of the mock-treated vector-only sample. Data shown represent the average from at least three independent transfection experiments. (B) Summary of the phenotypes of SV5 V or altered forms of SV5 V. The protein domains present in each form of SV5 V are shown on the left. Interaction with DDB1 was determined using the yeast two-hybrid assay; a strongly positive interaction (+++) was scored as the ability of transformants to give colonies in excess of 2 mm after 5 days' growth at 30°C on SDM lacking histidine plus 5 mM 3-aminotriazole and was comparable to that seen for the interaction between the SV40 T antigen and p53 (plasmids pVA3 and pTD1; Clontech). Colonies of 1 to 2 mm after 5 days are indicated by ++. No interaction (−) was indicated by colonies of less than 0.2 mm and was equivalent to the growth rate of the untransformed yeast strain or yeast transformed with control plasmid pGBT9 or pGBT9.SV5-V or doubly transformed with pGBT9.SV5-V and pHON3 (data not shown). Inhibition of IFN signaling was determined using the approach exemplified in panel A. Intact SV5 V and a truncation expressing only amino acids 20 to 222 limit IFN signaling to twofold or less (and are scored as +) in comparison to the 30- to 40-fold induction seen with vector alone. All other forms of SV5 V examined failed to inhibit IFN signaling and are therefore scored as −.
Direct evidence for the role of DDB1 in STAT1 degradation.
The observations that the loss of binding of V to DDB1 correlated with a loss in its ability to block IFN signaling and that the V protein of CPI− (which fails to target STAT1 for degradation) has a reduced affinity for DDB1 strongly suggested that there may be a central role for DDB1 in targeting STAT1 for proteasome-mediated degradation. To examine this more directly, we attempted to reduce the levels of DDB1 in 2f/SV5-V cells by treating the cells with siRNAs specific for DDB1. Although our preliminary experiments showed a reduction in the levels of DDB1, the effect was unconvincing (data not shown). We reasoned that this might be explained on the basis that 2fTGH cells are difficult to transfect, and therefore we repeated the siRNA experiments using a HeLa cell line which expressed the SV5 V protein [isolated as described earlier: HeLa/SV5-V(W3)]. Prior to siRNA treatment, STAT1 could not be detected either by immune blot analysis or by metabolically labeling the cells with [35S]methionine and immune precipitation (Fig. 4c and d, lanes 5 and 6). Encouragingly, treatment of the cells with siRNAs inhibited the synthesis of DDB1 (Fig. 4a). (Note that because the anti-DDB1 antibody does not work in the immune precipitation, DDB1 was coprecipitated with V using the anti-Pk MAb; consequently, DDB1 was not precipitated from naive HeLa cells.) Thus, while newly synthesized DDB1 was clearly visible in HeLa/SV5-V(W3) cells treated with control RNA (Fig. 4a, lanes 5 and 6), it could not be detected in cells treated with siRNAs (Fig. 4a, lanes 3 and 4). Furthermore, immunoblot analysis showed a clear, although not complete, reduction in the levels of DDB1 in siRNA-treated cells (Fig. 4b, lanes 3 and 4, compared to controls). In the same experiment, levels of STAT1 were also examined. In addition, since STAT1 is induced by IFN, the cells were or were not stimulated with IFN, since an increase in STAT1 levels would reflect functional IFN signaling. Strikingly, while STAT1 could not be immunoprecipitated from HeLa/SV5-V(W3) cells treated with control RNA (Fig. 4c, lanes 5 and 6), newly synthesized STAT1 was precipitated from cells that had been treated with siRNAs (Fig. 4c, lanes 3 and 4). However, the level of STAT1 in siRNA-treated cells was not as high as in control HeLa cells (Fig. 4c, compare lanes 1 and 3). Presumably, given the fact that DDB1 is not completely abolished by siRNA treatment, the kinetics of turnover of STAT1 are delayed but not completely abolished in siRNA-treated cells. Nevertheless, incubation of siRNA-treated cells with IFN significantly increased the levels of STAT1 (Fig. 4c, compare lanes 3 and 4). Indeed the increase of STAT1 in siRNA- and IFN-treated cells was such that STAT1 accumulated to levels that could be detected by immunoblot analysis (Fig. 4d, lane 4), demonstrating a restoration of IFN signaling in siRNA-treated cells. In contrast, incubation of IFN with HeLa/SV5-V(W3) cells that had been treated with control RNA had no effect on the detectable levels of STAT1 (Fig. 4c and d, compare lanes 5 and 6).
STAT1 is degraded in cells from a group E patient with xeroderma pigmentosum.
DDB has a high level of affinity for certain types of DNA lesions. It is a heterodimeric protein, consisting of two subunits, the 127-kDa DDB1 and a smaller 48-kDa subunit termed DDB2, which functions to activate DDB1 binding to damaged DNA. DDB1 is significantly more abundant than DDB2 (19, 41, 42) and moreover, unlike DDB2, is predominantly localized to the cytoplasm (29, 41), the presumptive site of STAT1 degradation. To investigate whether DNA binding activity was required for STAT1 degradation, the ability of SV5 to degrade STAT1 in GM02415 (2RO) cells, derived from a group E patient with xeroderma pigmentosum, was investigated. A single G-to-A transversion at nucleotide position 818 has been identified in the DDB2 gene of GM02415 cells which causes an R273H change. As a consequence, the R273H mutant DDB2 fails to associate with DDB1 (and cullin-4A; see Discussion) and is thus responsible for the absence of DDB activity in these cells (4, 31, 41). GM02415 cells were infected with SV5 and were or were not also treated with IFN from 0 to 24 h p.i. It can be seen from Fig. 5 that STAT1 was degraded in these cells, regardless of whether they had or had not been treated with IFN. Furthermore, in immune precipitation studies we have been unable to demonstrate an association between DDB2 and V (data not shown), suggesting that DDB2 is not involved in the degradation of STAT1 by SV5.
FIG. 5.
STAT1 is degraded in cells from a group E patient with xeroderma pigmentosum. GM02415 cells were infected with SV5 (strain W3) at a multiplicity of infection of 5. At the time of infection, the cells were (+) or were not (−) treated with IFN throughout the course of infection. At 24 h p.i. the cells were harvested, and the presence of STAT1 was detected by immunoblot analysis.
DISCUSSION
In an effort to understand the biochemical details of STAT targeting by paramyxoviruses and their specificity, we have begun a detailed analysis of the SV5-dependent process. Here we report that the interaction of V with DDB1 is required for the degradation of STAT1 by SV5, the most compelling evidence being that treatment of HeLa/SV5-V cells with siRNAs specific for DDB1 led to a reduction of DDB1 levels in treated cells with a concomitant increase the amount of STAT1 and a restoration of IFN signaling. Furthermore, given that SV5 degrades STAT1 in GMO2415 cells, the role of DDB1 in STAT1 degradation is independent of its association with DDB2. Since it has previously been shown that the hPIV2 V protein also interacts with DDB1 (27), it seems reasonable to presume that SV5 and hPIV2 utilize the same DDB1-dependent mechanism and the same downstream destruction pathway. We also present evidence that STAT2 is required for the degradation of STAT1 by SV5 and STAT1 is required for the degradation of STAT2 by hPIV2. These last results are therefore in agreement with those recently published by Parisien and colleagues (35), who came to a similar conclusion by complementing STAT-deficient cell lines. It therefore seems likely that during the degradation of STAT1 and STAT2 by SV5 or hPIV2, respectively, a large multiprotein complex may be formed in which V, STAT1, STAT2, and DDB1 are essential components. While such a complex has yet to be isolated, it has been reported that as well as binding DDB1, the V protein of SV5 can bind, directly or indirectly, both STAT1 and STAT2 (35). The involvement of a stoichiometric STAT1/STAT2 complex is further strengthened by our observation that in transient transfections assays where STAT1 is overexpressed, degradation was seen only if STAT2 was also overexpressed. However, unlike the phosphotyrosine-dependent heterodimerization of STAT1 and STAT2 seen in response to IFN, the putative ligand independent complex induced by V would be formed in cells not induced by IFN, since neither active IFN signaling nor the phosphorylation of STAT1 or STAT2 is required for STAT degradation by either SV5 or hPIV2 (1, 35). One intriguing aspect of this work is the selective targeting of STAT1 or STAT2 for degradation by SV5 or hPIV2, respectively. In a model in which the viral V proteins recruit STAT1/STAT2 heterodimers to DDB1, it is difficult to see why only one of the STAT partners gets degraded and why the identity of this partner should differ. If so, then the selective targeting of STAT1 or STAT2 is likely to be a consequence of the relative conformation of the STAT1/STAT2 heterodimer as presented to the degradative machinery, which would differ between SV5 and hPIV2.
It has been previously shown that the binding of V to DDB1 is dependent upon the cysteine-rich C-terminal unique domain of V. Data presented here not only confirm this requirement but extend the findings to demonstrate that residues in the N-terminal P/V common domain also influence the binding of V to DDB1. Thus, only relatively short N-terminal truncations of V can be tolerated before the binding of V to DDB1 is abolished. Furthermore, the C-unique terminus of CPI− is also identical to that of W3 and CPI+, and yet CPI− fails to bind DDB1 in the assays employed. Indeed, there are only three amino acid differences between the V proteins of CPI+ and CPI−, and all of these are located in the N terminus. (It is possible that the reduced affinity of the V protein of CPI− for DDB1 is the sole reason why CPI− fails to target STAT1 for degradation.) However, since the P protein of SV5 does not bind DDB1, it seems unlikely that the N terminus P/V common domain alone can bind DDB1. Whether DDB1 binds to sequences throughout V or whether the mutations in the N terminus of V alter the structure of the carboxy terminus may not be resolved until the three-dimensional structure of V has been resolved.
In addition to binding the V proteins of many paramyxoviruses, DDB1 has been shown to bind to the Hepatitis B virus X protein (28, 43, 44, 51), the cytoplasmic tail of the amyloid protein precursor (50), and cullin-4A (4, 31, 42). The fact that cullins, as part of ubiquitin E3 ligases (16, 33, 45; for a review, see reference 38), play an essential role in targeting proteins for degradation through proteasomes points to a possible role for cullin 4A in the degradation of STAT1. An attractive model, which we are currently exploring, is that the V protein of SV5 acts as a bridge to bring STAT1/STAT2 together with DDB1, which then binds to cullin-4A in a ubiquitin E3 ligase complex, resulting ubiquitination and degradation of STAT1. Although cullin-4A stimulates the ubiquitination of DDB1 (4), there is no significant change to the overall levels of DDB1, raising the possibility that DDB1 may play a chaperone role, i.e., it recruits the viral V protein and, indirectly, the STAT proteins to the ubiquitin E3 ligase, without DDB1 necessarily being degraded. However, it is also becoming clear that there are a great variety of ubiquitin E3 ligases beyond the prototypical SCF complex (38), and thus, DDB1 may form an integral part of a currently unidentified ubiquitin E3 ligase involved in the degradation of STAT1/STAT2. Furthermore, although we have shown that the degradation of STAT1 during SV5 infections can be blocked by the proteasome inhibitors MG132 and lactacystin (7), so far we have failed to detect polyubiquitinylated forms of STAT1. It is thus also possible that STAT1 degradation in response to viral V protein may be independent of ubiquitinylation. This is not unprecedented; for example, adenovirus E1A proteasome-mediated degradation is not mediated through ubiquitinylation (49). Thus, by elucidating the molecular basis of STAT degradation by the V proteins of SV5 and hPIV2, not only will insights into virus host cell pathogenesis be gained but also the normal cellular functions of proteins, such as DDB1, may be further understood.
Acknowledgments
R. A. Lamb (Northwestern University, Evanston, Ill.) supplied the V mAb 11, and V. Rapic-Otrin (University of Pittsburgh, Pittsburgh, Pa.) provided us with the anti-DDB1 antibody and the plasmid pCEPp127, for which we are extremely grateful.
J. Andrejeva, E. Poole, and D. Young are supported by grants from the Wellcome Trust and BBSRC.
REFERENCES
- 1.Andrejeva, J., D. F. Young, S. Goodbourn, and R. E. Randall. 2002. Degradation of STAT1 and STAT2 by the V proteins of simian virus 5 and human parainfluenza virus type 2, respectively: consequences for virus replication in the presence of alpha/beta and gamma interferons. J. Virol. 76:2159-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel, P. L., C.-T. Chien, R. Sternglanz and S. Fields. 1993. Using the two-hybrid system to detect protein-protein interactions, p. 153-179. Cellular interactions in development: a practical approach. Oxford University Press, Oxford, United Kingdom.
- 3.Chatziandreou, N., D. Young, J. Andrejeva, S. Goodbourn, and R. E. Randall. 2002. Differences in interferon sensitivity and biological properties of two related isolates of simian virus 5: a model for virus persistence. Virology 293:234-242. [DOI] [PubMed] [Google Scholar]
- 4.Chen, X., Y. Zhang, L. Douglas, and P. Zhou. 2001. UV-damaged DNA-binding proteins are targets of CUL-4A-mediated ubiquitination and degradation. J. Biol. Chem. 276:48175-48182. [DOI] [PubMed] [Google Scholar]
- 5.Choppin, P. W. 1964. Multiplication of a myxovirus (SV5) with minimal cytopathic effects and without interference. Virology 23:224-233. [DOI] [PubMed] [Google Scholar]
- 6.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. Sendai virus and simian virus 5 block activation of interferon-responsive genes: importance for virus pathogenesis. J. Virol. 73:3125-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. The V protein of simian virus 5 inhibits interferon signalling by targeting STAT1 for proteasome-mediated degradation. J. Virol. 73:9928-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 9.Garcin, D., J. Curran, and D. Kolakofsky. 2000. Sendai virus C proteins must interact directly with cellular components to interfere with interferon action. J. Virol. 74:8823-8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcin, D., P. Latorre, and D. Kolakofsky. 1999. Sendai virus C proteins counteract the interferon-mediated induction of an antiviral state. J. Virol. 73:6559-6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gietz, R. D., R. H. Schiestl, A. R. Willems, and R. A. Woods. 1995. Studies on the transformation of intact yeast cells by the LiAc/SS- DNA/PEG procedure. Yeast 11:355-360. [DOI] [PubMed] [Google Scholar]
- 12.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 13.Gotoh, B., K. Takeuchi, T. Komatsu, J. Yokoo, Y. Kimura, A. Kurotani, A. Kato, and Y. Nagai. 1999. Knockout of the Sendai virus C gene eliminates the viral ability to prevent the interferon-alpha/beta-mediated responses. FEBS Lett. 459:205-210. [DOI] [PubMed] [Google Scholar]
- 14.Itoh, T., S. Linn, T. Ono, and M. Yamaizumi. 2000. Reinvestigation of the classification of five cell strains of xeroderma pigmentosum group E with reclassification of three of them. J. Investig. Dermatol. 114:1022-1029. [DOI] [PubMed] [Google Scholar]
- 15.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamura, T., D. M. Koepp, M. N. Conrad, D. Skowyra, R. J. Moreland, O. Iliopoulos, W. S. Lane, W. G. Kaelin, Jr., S. J. Elledge, R. C. Conaway, J. W. Harper, and J. W. Conaway. 1999. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science 284:657-661. [DOI] [PubMed] [Google Scholar]
- 17.Kato, A., Y. Ohnishi, M. Kohase, S. Saito, M. Tashiro, and Y. Nagai. 2001. Y2, the smallest of the Sendai virus C proteins, is fully capable of both counteracting the antiviral action of interferons and inhibiting viral RNA synthesis. J. Virol. 75:3802-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawano, M., M. Kaito, Y. Kozuka, H. Komada, N. Noda, K. Nanba, M. Tsurudome, M. Ito, M. Nishio, and Y. Ito. 2001. Recovery of infectious human parainfluenza type 2 virus from cDNA clones and properties of the defective virus without V-specific cysteine-rich domain. Virology 284:99-112. [DOI] [PubMed] [Google Scholar]
- 19.Keeney, S., G. J. Chang, and S. Linn. 1993. Characterization of a human DNA damage binding protein implicated in xeroderma pigmentosum E. J. Biol. Chem. 268:21293-21300. [PubMed] [Google Scholar]
- 20.King, P., and S. Goodbourn. 1994. The beta-interferon promoter responds to priming through multiple independent regulatory elements. J. Biol. Chem. 269:30609-30615. [PubMed] [Google Scholar]
- 21.King, P., and S. Goodbourn. 1998. STAT1 is inactivated by a caspase. J. Biol. Chem. 273:8699-8704. [DOI] [PubMed] [Google Scholar]
- 22.Komatsu, T., K. Takeuchi, J. Yokoo, and B. Gotoh. 2002. Sendai virus C protein impairs both phosphorylation and dephosphorylation processes of Stat1. FEBS Lett. 511:139-144. [DOI] [PubMed] [Google Scholar]
- 23.Komatsu, T., K. Takeuchi, J. Yokoo, Y. Tanaka, and B. Gotoh. 2000. Sendai virus blocks alpha interferon signaling to signal transducers and activators of transcription. J. Virol. 74:2477-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kubota, T., N. Yokosawa, S. Yokota, and N. Fujii. 2001. C terminal CYS-RICH region of mumps virus structural V protein correlates with block of interferon alpha and gamma signal transduction pathway through decrease of STAT 1-alpha. Biochem. Biophys. Res. Commun. 283:255-259. [DOI] [PubMed] [Google Scholar]
- 25.Levy, D. E., and A. Garcia-Sastre. 2001. The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine Growth Factor Rev. 12:143-156. [DOI] [PubMed] [Google Scholar]
- 26.Lin, G. Y., and R. A. Lamb. 2000. The paramyxovirus simian virus 5 V protein slows progression of the cell cycle. J. Virol. 74:9152-9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin, G. Y., R. G. Paterson, C. D. Richardson, and R. A. Lamb. 1998. The V protein of the paramyxovirus SV5 interacts with damage-specific DNA binding protein. Virology 249:189-200. [DOI] [PubMed] [Google Scholar]
- 28.Lin-Marq, N., S. Bontron, O. Leupin, and M. Strubin. 2001. Hepatitis B virus X protein interferes with cell viability through interaction with the p127-kDa UV-damaged DNA-binding protein. Virology 287:266-274. [DOI] [PubMed] [Google Scholar]
- 29.Liu, W., A. F. Nichols, J. A. Graham, R. Dualan, A. Abbas, and S. Linn. 2000. Nuclear transport of human DDB protein induced by ultraviolet light. J. Biol. Chem. 275:21429-21434. [DOI] [PubMed] [Google Scholar]
- 30.McKendry, R., J. John, D. Flavell, M. Muller, I. M. Kerr, and G. R. Stark. 1991. High-frequency mutagenesis of human cells and characterization of a mutant unresponsive to both alpha and gamma interferons. Proc. Natl. Acad. Sci. USA 88:11455-11459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nag, A., T. Bondar, S. Shiv, and P. Raychaudhuri. 2001. The xeroderma pigmentosum group E gene product DDB2 is a specific target of cullin 4A in mammalian cells. Mol. Cell. Biol. 21:6738-6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishio, M., M. Tsurudome, M. Ito, M. Kawano, H. Komada, and Y. Ito. 2001. High resistance of human parainfluenza type 2 virus protein-expressing cells to the antiviral and anti-cell proliferative activities of alpha/beta interferons: cysteine-rich V-specific domain is required for high resistance to the interferons. J. Virol. 75:9165-9176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohta, T., J. J. Michel, A. J. Schottelius, and Y. Xiong. 1999. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol. Cell 3:535-541. [DOI] [PubMed] [Google Scholar]
- 34.Parisien, J. P., J. F. Lau, J. J. Rodriguez, B. M. Sullivan, A. Moscona, G. D. Parks, R. A. Lamb, and C. M. Horvath. 2001. The V protein of human parainfluenza virus 2 antagonizes type I interferon responses by destabilizing signal transducer and activator of transcription 2. Virology 283:230-239. [DOI] [PubMed] [Google Scholar]
- 35.Parisien, J. P., J. F. Lau, J. J. Rodriguez, C. M. Ulane, and C. M. Horvath. 2002. Selective STAT protein degradation induced by paramyxoviruses requires both STAT1 and STAT2 but is independent of alpha/beta interferon signal transduction. J. Virol. 76:4190-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paterson, R. G., G. P. Leser, M. A. Shaughnessy, and R. A. Lamb. 1995. The paramyxovirus SV5 V protein binds two atoms of zinc and is a structural component of virions. Virology 208:121-131. [DOI] [PubMed] [Google Scholar]
- 37.Pellegrini, S., J. John, M. Shearer, I. M. Kerr, and G. R. Stark. 1989. Use of a selectable marker regulated by alpha interferon to obtain mutations in the signaling pathway. Mol. Cell. Biol. 9:4605-4612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pickart, C. M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70:503-533. [DOI] [PubMed] [Google Scholar]
- 39.Randall, R. E., and A. Bermingham. 1996. NP:P and NP:V interactions of the paramyxovirus simian virus 5 examined using a novel protein:protein capture assay. Virology 224:121-129. [DOI] [PubMed] [Google Scholar]
- 40.Saito, S., T. Ogino, N. Miyajima, A. Kato, and M. Kohase. 2002. Dephosphorylation failure of tyrosine-phosphorylated STAT1 in IFN-stimulated Sendai virus C protein-expressing cells. Virology 293:205-209. [DOI] [PubMed] [Google Scholar]
- 41.Shiyanov, P., S. A. Hayes, M. Donepudi, A. F. Nichols, S. Linn, B. L. Slagle, and P. Raychaudhuri. 1999. The naturally occurring mutants of DDB are impaired in stimulating nuclear import of the p125 subunit and E2F1-activated transcription. Mol. Cell. Biol. 19:4935-4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shiyanov, P., A. Nag, and P. Raychaudhuri. 1999. Cullin 4A associates with the UV-damaged DNA-binding protein DDB. J. Biol. Chem. 274:35309-35312. [DOI] [PubMed] [Google Scholar]
- 43.Sitterlin, D., F. Bergametti, P. Tiollais, B. C. Tennant, and C. Transy. 2000. Correct binding of viral X protein to UVDDB-p127 cellular protein is critical for efficient infection by hepatitis B viruses. Oncogene 19:4427-4431. [DOI] [PubMed] [Google Scholar]
- 44.Sitterlin, D., F. Bergametti, and C. Transy. 2000. UVDDB p127-binding modulates activities and intracellular distribution of hepatitis B virus X protein. Oncogene 19:4417-4426. [DOI] [PubMed] [Google Scholar]
- 45.Skowyra, D., D. M. Koepp, T. Kamura, M. N. Conrad, R. C. Conaway, J. W. Conaway, S. J. Elledge, and J. W. Harper. 1999. Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science 284:662-665. [DOI] [PubMed] [Google Scholar]
- 46.Southern, J. A., D. F. Young, F. Heaney, W. K. Baumgartner, and R. E. Randall. 1991. Identification of an epitope on the P and V proteins of simian virus 5 that distinguishes between two isolates with different biological characteristics. J. Gen. Virol. 72:1551-1557. [DOI] [PubMed] [Google Scholar]
- 47.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 48.Takeuchi, K., T. Komatsu, J. Yokoo, A. Kato, T. Shioda, Y. Nagai, and B. Gotoh. 2001. Sendai virus C protein physically associates with Stat1. Genes Cells 6:545-557. [DOI] [PubMed] [Google Scholar]
- 49.Turnell, A. S., R. J. Grand, C. Gorbea, X. Zhang, W. Wang, J. S. Mymryk, and P. H. Gallimore. 2000. Regulation of the 26S proteasome by adenovirus E1A. EMBO J. 19:4759-4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Watanabe, T., J. Sukegawa, I. Sukegawa, S. Tomita, K. Iijima, S. Oguchi, T. Suzuki, A. C. Nairn, and P. Greengard. 1999. A 127-kDa protein (UV-DDB) binds to the cytoplasmic domain of the Alzheimer's amyloid precursor protein. J. Neurochem. 72:549-556. [DOI] [PubMed] [Google Scholar]
- 51.Wentz, M. J., S. A. Becker, and B. L. Slagle. 2000. Dissociation of DDB1-binding and transactivation properties of the hepatitis B virus X protein. Virus Res. 68:87-92. [DOI] [PubMed] [Google Scholar]
- 52.Young, D. F., N. Chatziandreou, B. He, S. Goodbourn, R. A. Lamb, and R. E. Randall. 2001. Single amino acid substitution in the V protein of simian virus 5 differentiates its ability to block interferon signaling in human and murine cells. J. Virol. 75:3363-3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Young, D. F., L. Didcock, S. Goodbourn, and R. E. Randall. 2000. Paramyxoviridae use distinct virus-specific mechanisms to circumvent the interferon response. Virology 269:383-390. [DOI] [PubMed] [Google Scholar]