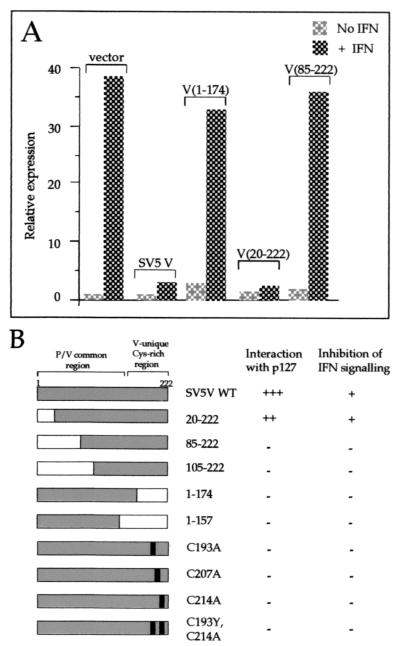
FIG. 3.
(A) Examples of the ability of altered SV5 V proteins to block IFN signaling are shown. HeLa cells were transfected with plasmids as indicated, a luciferase reporter plasmid, and a β-galactosidase expression vector. Cells were or were not treated with 1,000 U of IFN-á for 4 h; luciferase levels in extracts were normalized to β-galactosidase levels, and the relative expression values were plotted; a value of 1.0 was assigned to the basal expression level of the mock-treated vector-only sample. Data shown represent the average from at least three independent transfection experiments. (B) Summary of the phenotypes of SV5 V or altered forms of SV5 V. The protein domains present in each form of SV5 V are shown on the left. Interaction with DDB1 was determined using the yeast two-hybrid assay; a strongly positive interaction (+++) was scored as the ability of transformants to give colonies in excess of 2 mm after 5 days' growth at 30°C on SDM lacking histidine plus 5 mM 3-aminotriazole and was comparable to that seen for the interaction between the SV40 T antigen and p53 (plasmids pVA3 and pTD1; Clontech). Colonies of 1 to 2 mm after 5 days are indicated by ++. No interaction (−) was indicated by colonies of less than 0.2 mm and was equivalent to the growth rate of the untransformed yeast strain or yeast transformed with control plasmid pGBT9 or pGBT9.SV5-V or doubly transformed with pGBT9.SV5-V and pHON3 (data not shown). Inhibition of IFN signaling was determined using the approach exemplified in panel A. Intact SV5 V and a truncation expressing only amino acids 20 to 222 limit IFN signaling to twofold or less (and are scored as +) in comparison to the 30- to 40-fold induction seen with vector alone. All other forms of SV5 V examined failed to inhibit IFN signaling and are therefore scored as −.