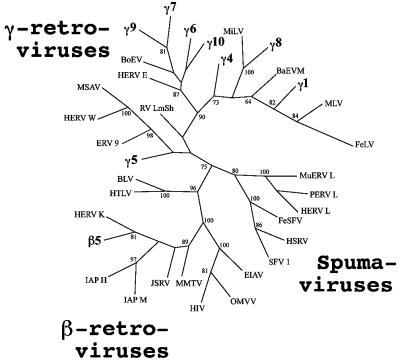
FIG. 1.
Neighbor-joining tree of retroviruses produced by PHYLIP. The analysis was done with the 510-nt 3′ fragment of the amplified pro-pol sequences and a data set of 1,000 bootstrap replicates. Percent bootstrap values higher than 60% are shown. The PERV sequences obtained in this study (γ1, γ4 to γ10, and β5) are depicted in bold. The viruses and GenBank accession numbers of the sequences (in parentheses) used in the phylogenetic tree are as follows: BaEVM (M16550), baboon ERV; BLV (AF257515), bovine leukemia virus; BoEV (X99924), bovine ERV; EIAV (AF247394), equine infectious anemia virus; ERV 9 (X57147), human ERV 9; FeLV (AF052723), feline leukemia virus; FeSFV (U78765), feline syncytial virus; HERV E (M10976), human ERV E; HERV K (M14123); HERV L (X89211); HERV W (AF135487); HIV (K03455), human immunodeficiency virus; HSRV (U21247), human spumaretrovirus; (L03561), human T-cell lymphotropic virus; IAP H (M10134), syrian hamster intracisternal A-particle; IAP M (M17551), murine intracisternal A-particle; JSRV (M80216), Jaagsiekte sheep retrovirus; MiLV (X99931), mink ERV; MLV (J01998), murine leukemia virus; MMTV (M15122), mouse mammary tumor virus; MSAV (AF009668), multiple sclerosis-associated retrovirus; MuERV L (Y12713), murine ERV L; OMVV (NC_001511), ovine lentivirus; PERV L (AJ233661); RVLmSh (Y07810), lemon shark ERV; and SFV 1 (X54482), simian foamy virus 1.