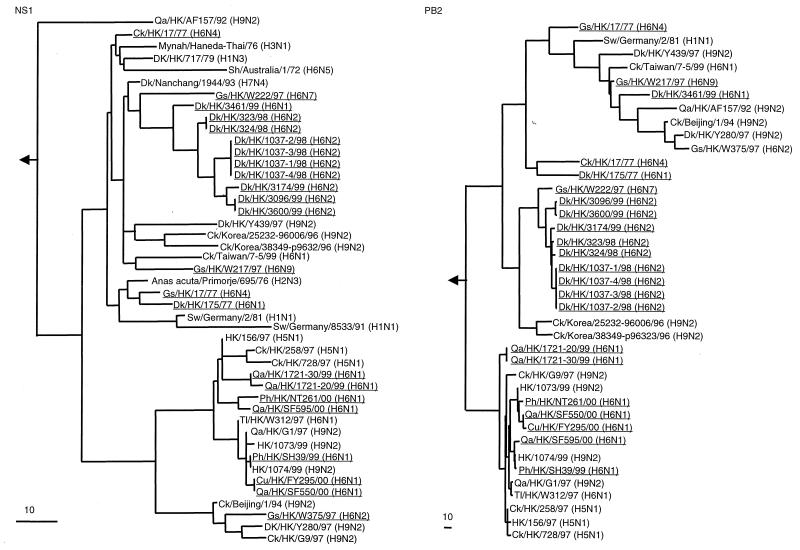
FIG. 2.
Phylogenetic trees based on the sequences of the NS1 and PB2 genes of influenza viruses. H6 influenza viruses characterized in this study are underlined. The ranges of nucleotide sequences used to construct the phylogenetic trees are shown in Table 4. The NS1 phylogenetic tree is rooted to A/equine/Prague/1/56 (H7N7), and the PB2 phylogenetic tree is rooted to B/Lee/40. The lengths of the horizontal lines are proportional to the minimum number of nucleotide differences required to join nodes. Vertical lines are for spacing branches and labels. Gene sequences used in this study are listed under the following EMBL Nucleotide Sequence Database accession numbers: AF156479, M17070, M60800, M55484, Z26865, U49492, L25830, AF156476, AF262212, U49493, AF250502, AF156482, AF156481, AF036360, AF098569, AF098571, AF250483, AF156477, AJ278649, AJ404735, AF156480, AF156472, AF156475, AF156440, AF156439, M55471, AF255624, AF156434, AF156437, AF156438, AF156436, AF156433, AF156430, AJ404630, AJ404630, AF036363, AF098577, AF098579, AF250476, and AF156435. Sw, swine.