Figure 1.
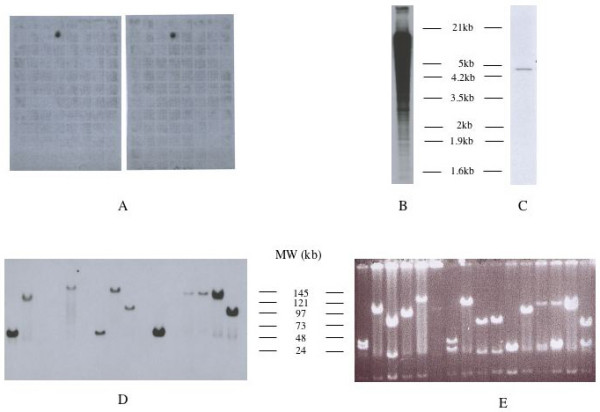
Isolation and characterization of BACS. Panel A shows typical results obtained with a BAC containing a MAV-integration site contained in one of the probes dereived from avian nephroblastoma. Filters of BAC DNA were duplicated. To check the specificity of the probes used, two micrograms of genomic chicken DNA were digested with 40 units of HindIII restriction endonuclease at 37°C for 18 hours and run in a 1% agarose gel at 2 volts/cm for 20 hours. The separated DNA fragments were denatured by incubation in 0.5 N NaOH for 45 min, neutralized in 0.5 M Tris HCl, 1.5 M NaCl. Transfer onto Appligene Positive Membrane was performed in 20 × SSC for 18 hours and the membrane was baked for 2 hours at 80°C prior to use for hybridization with labeled cloned cellular fragments. All cellular probes cloned from nephroblastoma DNA libraries detected a single fragment in HinIII digested normal DNA (see panel C for typical result) except for P38 which contained chicken repetitive sequences (panel B). Panels D and E: DNA preparations from positive Bacs were digested with NotI (panel 1 E shows ethidium bromide staining of the gel) and transfered onto nitrocellulose prior to hybridization with the probes used for their isolation. A single NotI fragment is detected by the probes in the BAC dans (panel 1D).
