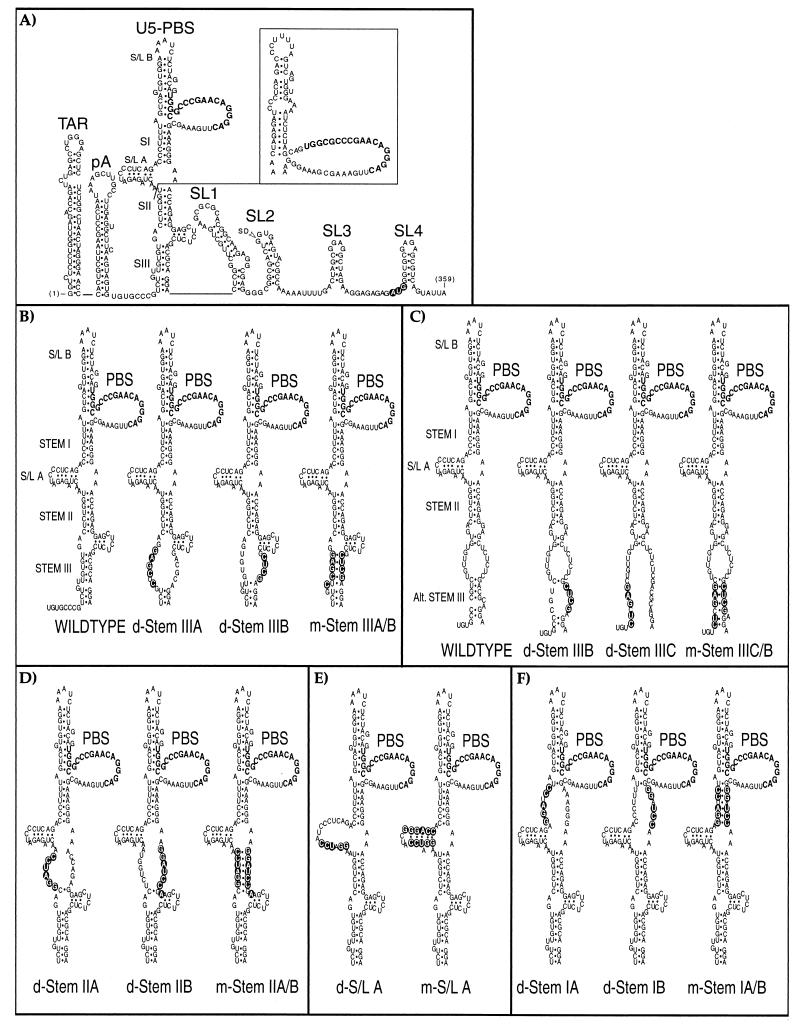
FIG.1.
Mutant HIV-1 RNAs characterized in this study. (A) The 5′ leader region of wild-type HIV-1 RNA from strain HXB2, with the gag initiation codon shown with open lettering and the major 5′ splice donor (SD) indicated. The conformations shown for the TAR, poly(A) (pA), and SL1 to SL4 stem-loops were extensively validated by earlier genetic, functional, and structural data. The U5-PBS region, which is the focus of this study, is shown in two proposed conformations that are compatible with the results of published accessibility mapping studies (2, 15). The three major postulated stems (SI, SII, and SIII) are identified as described previously (2); two associated stem-loop elements are designated S/L A and S/L B in this study, and the 18 nucleotides (nt) of the PBS are shown in boldface type. An alternative conformation (15) for the distal sequences is shown as an insert. (B to F) The sequences and putative structural consequences of the 13 mutations tested in this study. Mutations predicted to disrupt base pairing are designated with the prefix “d-” whereas compensatory mutations designed to preserve potential base pairing have “m-” as the prefix. In most cases, compensatory mutations were constructed by combining a pair of disruptive mutations. The mutations shown in panels B and C were designed to test two alternative conformations of stem III. BspEI was used to linearize the plasmid DNA before the riboprobes used in the RNase protection assays were synthesized. An additional nucleotide outside stem II was altered in d-Stem IIB and m-Stem IIA/B to avoid creating a second BspEI restriction enzyme site; for the same reason, one nucleotide outside the stem of S/L A was altered in the m-S/L A mutant.