Figure 3.
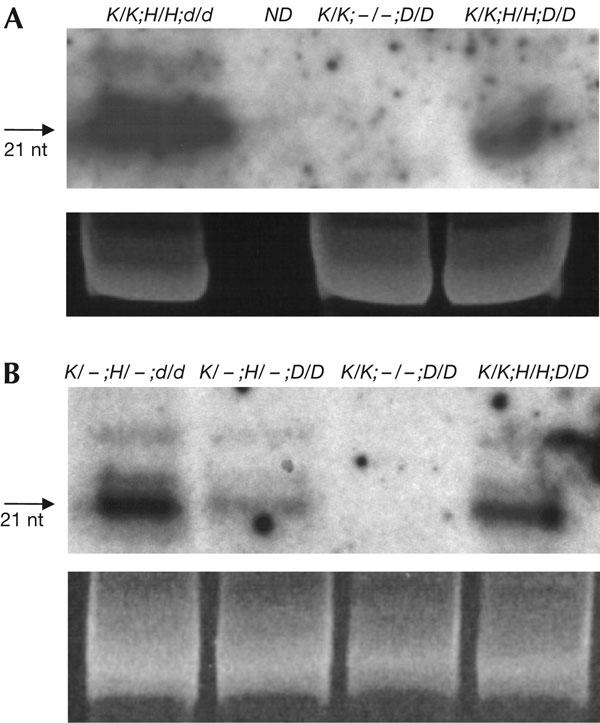
Short RNA analysis. (A) Northern blot analysis of α′ promoter short RNAs in wild-type plants containing only the target complex (K/K;−/−;D/D) or both the target and silencer complexes (K/K;H/H;D/D), or in the drd1-6 mutant (K/K;H/H;d/d). (B) Northern blot analysis of nopaline synthase (NOS) promoter short RNAs in F1 progeny from the ‘de novo' cross (Fig 2) that were homozygous for the drd1-6 mutation (K/−;H/−;d/d) or wild type (K/−;H/−;D/D), and in wild-type plants that were either homozygous for the target K complex only (K/K;−/−;D/D) or double homozygous for the target K complex and silencer H complex (K/K;H/H;D/D). In wild-type plants, NOS promoter short RNAs are approximately twice as abundant in homozygous H/H as in hemizygous H/− plants. The quantities of short RNAs are increased in drd1-6 mutants, perhaps because they are not used and turned over efficiently in these plants. Ethidium bromide staining of the principal RNA species in the samples is shown as a loading control. ND indicates that α′ promoter short RNAs were not tested in the genetype K/−;H/−;D/D.
