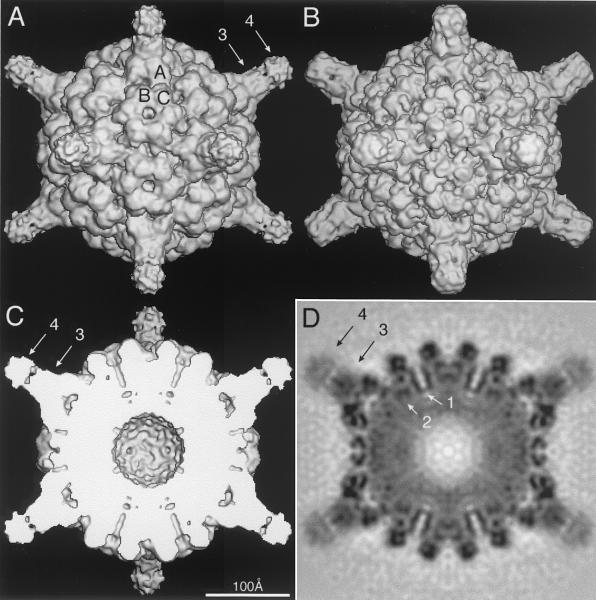
FIG. 2.
Shaded-surface (A to C) and equatorial section (D) representations of the CMV-Fab complex, all viewed along a twofold axis of symmetry. The arrows highlight aspects of the cryoTEM 3D reconstruction that are depicted in greater detail in subsequent figures. (A) 3D reconstruction of the CMV-Fab complex with the approximate locations of the three quasiequivalent (chemically identical) A, B, and C subunits labeled. The large, turret-like projections at the icosahedral vertices represent the bound Fab molecules, and the variable and constant domains of one Fab are identified by arrows 3 and 4, respectively. (B) Pseudo-atomic model of CMV-Fab complex computed to 10-Å resolution; β factors were applied to match the 12-Å cryo-TEM data, using the X-ray crystal structures of CMV (32) and Fab17 (19). (C) Same as panel A with the front half of the structure removed to visualize internal features of the reconstruction. (D) Density projection of the central section of the CMV-Fab reconstruction, with the blackest regions representing the highest density regions in the map. Two key internal features are the radially directed density representing the six-helical bundle at the quasi-sixfold axes (arrow 1) and density that lies adjacent to and projects away from the helical bundle (arrow 2). The lower density for the Fabs (especially the constant domains [arrow 4]) is consistent with the hypothesis that only a single Fab binds to each of the 12 pentons at the vertices of the icosahedral virus.