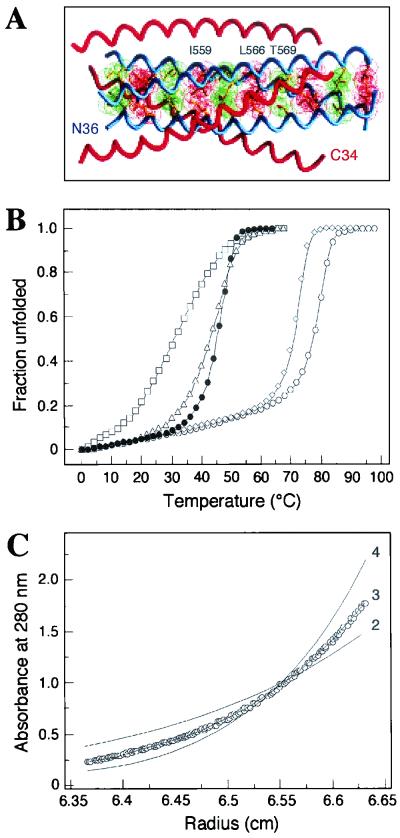
FIG. 6.
HIV-1 gp41ECTO core structure and mutant peptides. (A) The N36-C34 crystal structure is shown with one N36 helix and one C34 helix labeled at the amino termini. Three C34 helices (red) pack against the N36 trimeric coiled coil (blue). The van der Waal surfaces of residues at positions a (red) and d (green) are superimposed on the helix backbone of the N36 coiled coil. Amino acids substituted in this study are indicated above the a and d layers. The diagram was prepared by using the program GRASP (64). (B) Thermal melting transition curves of the N36(L6)C34 (open circles), I559G (closed circles), I559P (open squares), L566V (open diamonds), and T569P (open triangles) peptides were determined by CD spectroscopy at 222 nm and at a peptide concentration of 10 μM in PBS. The increase in the fraction of unfolded molecules is shown as a function of temperature. All melts were reversible. Superimposable folding and unfolding curves were observed, and >90% of the signal was regained upon cooling. (C) Equilibrium sedimentation analysis of the T569P peptide.Representative data for this peptide were collected at 20°C, 20,000 rpm, and a peptide concentration of ∼30 μM in PBS. The data fit best to a trimer model (curve 3). Curves for a dimer (curve 2) and a tetramer (curve 4) are depicted for comparison. Analyses of residual differences from curve 3 did not reveal a systematic error.