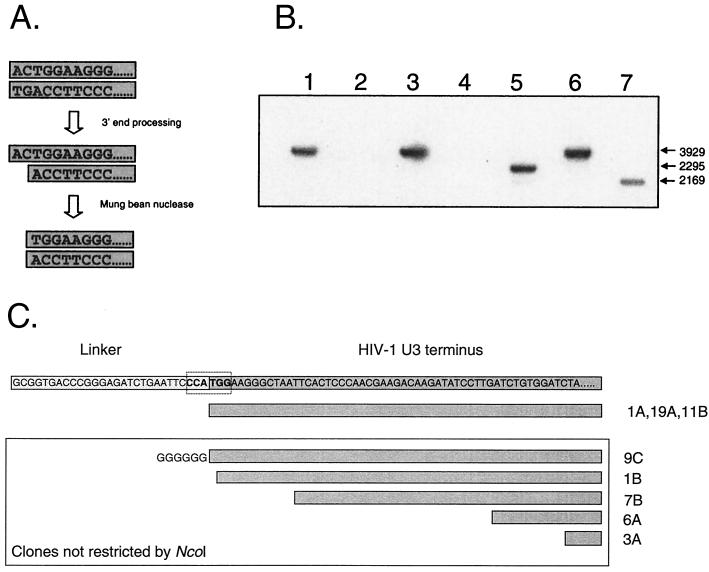
FIG. 5.
Infected resting CD4+ T cells contain linear HIV-1 DNA molecules that have 3′ processing by integrase. (A) LM-PCR assay for complete reverse transcripts that have undergone 3′ processing by integrase. As an initial step in the integration reaction, integrase removes the terminal 2 nucleotides from the 3′ end of each strand of the full-length, linear HIV-1 DNA. The product of this reaction can be detected by removing the 5′ overhang with single-strand-specific MBN, generating a blunt-ended molecule whose terminal nucleotides form one-half of an NcoI site. The linker used in these experiments creates a novel NcoI site when ligated to the linear genome of HIV-1 that had been processed by integrase and cleaved by MBN. LM-PCR clones were restricted with NcoI, run on a 1% gel, and probed by Southern blotting with an LTR-specific probe. The vector used in these studies has an NcoI site; therefore, clones with inserts derived from linear DNA that has been processed by integrase will be cleaved twice in a manner analogous to that for the ScaI screen described in the legend for Fig. 4. (B) LM-PCR analysis of HIV-1 DNA from infected resting CD4+ T cells. Highly purified resting CD4+ T cells were infected with HIV-1 IIIB. Three days after infection, cells were lysed and DNA was isolated for LM-PCR. Restriction analysis of cloned LM-PCR products identified cloned inserts that were cleaved by NcoI, indicating prior processing by integrase (lanes 5 and 7) and inserts that were not cleaved (lanes 1, 3, and 6). Because the insert was cloned in either a forward or reverse orientation, inserts of two different sizes were predicted for clones containing the novel NcoI site. Some clones did not contain an HIV-1-derived insert (lanes 2 and 4). (C) Sequence analysis of LM-PCR clones obtained from MBN-treated DNA isolated from infected resting T lymphocytes.