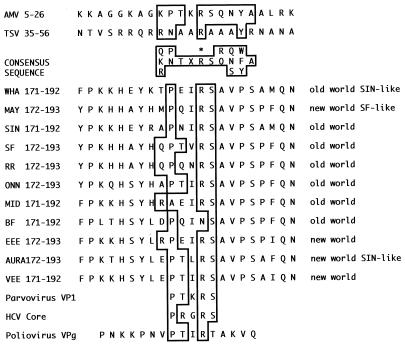
FIG. 1.
Amino acid alignment of the Pro-X-X-Arg-Ser sequence. Numbers denote the amino acid positions in the nsP4 proteins of the alphaviruses or for avian myeloblastosis virus (AMV) and tobacco streak virus (TSV) coat protein consensus sequence Pro-Thr-X-Arg-Ser (48). The substitution in SIN nsP4 conferring ts reactivation of minus-strand synthesis (34) is Gln191→Lys (34), and that conferring a host-restricted, ts RNA-negative phenotype in 35.1C2a virus (22) is Pro187→Arg (+). The consensus sequence determined for AMV and ilarvirus (1) coat protein is boxed, and the essential Arg (R∗) is indicated. References for the sequences are as follows: Whatora virus (WHA), Ellen Strauss, personal communication; Mayora virus (MAY), Ellen Strauss, personal communication; SIN, 46; SFV, 49; Ross River virus (RR), 8; O'nyong-nyong virus (ONN), 26; Middleburg virus (MID), 46; eastern equine encephalitis virus (EEE), 53; Aura virus, 33; Venezuelan equine encephalitis virus (VEE), 16; Barmah Forest virus (BF), 20; parvovirus VP1 and hepatitis C virus (HCV) core, 48; and poliovirus VPg, 30.