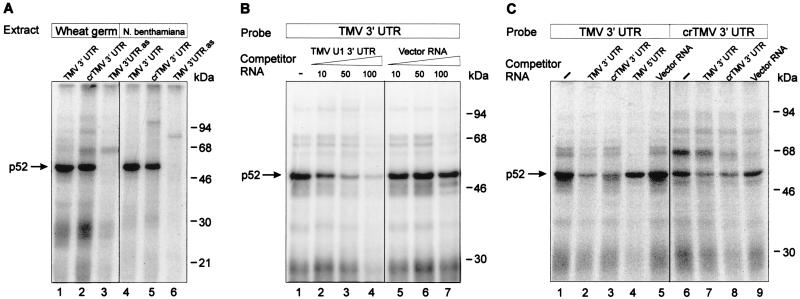
FIG. 2.
Specific binding of p52 to the 3′-UTRs of TMV U1 and crTMV RNAs. (A) Binding of proteins to the 3′-UTRs of TMV U1 and crTMV RNAs. UV light induced cross-linking of wheat germ (lanes 1 to 3) or N. benthamiana (lanes 4 to 6) S-100 extract proteins to 32P-labeled RNA probes as indicated above the lanes. Binding reactions were performed as described in Materials and Methods. The complexes formed were irradiated with UV light and then treated with RNases and analyzed by SDS-12% PAGE followed by autoradiography. The major cross-linked protein (p52) is indicated by an arrow. The positions of size markers are shown on the right. An RNA complementary to the TMV U1 3′-UTR (TMV 3′ UTR.as) was used as the control RNA in lanes 3 and 6. (B) Specificity of p52 binding to the TMV U1 3′-UTR. Binding reactions were carried out with wheat germ extract with 30 fmol (100,000 cpm) of 32P-labeled TMV U1 3′-UTR in the absence (lane 1) or in the presence of increasing amounts of unlabeled specific (lanes 2 to 4) and nonspecific (lanes 5 to 7) RNA competitors. The fold molar excess of competitor is indicated above each lane . (C) Cross-competition of the TMV U1 3′-UTRs, crTMV 3′-UTR, the TMV 5′-UTR and nonviral RNA. Conditions of binding were as described for panel A, except that a 100-fold molar excess of competitor was added in each case.