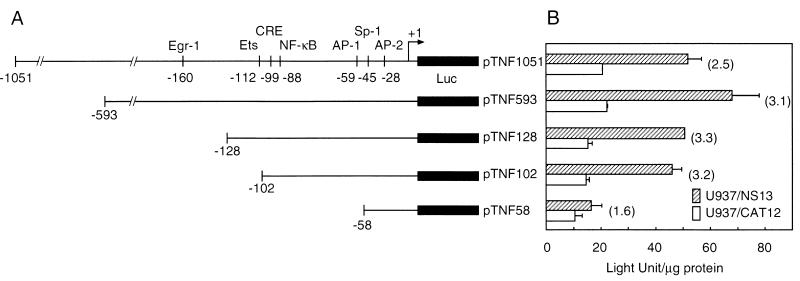
FIG. 3.
Effects of deletion mutations on the transcription activity of the TNF-α promoter in response to NS1 expression. (A) Schematic presentation of the TNF-α promoter region and its 5′ truncated forms. Putative cis-acting regulatory motifs in the reporter constructs are denoted, and the numbers identify the position relative to the transcription initiation site. Luc, luciferase. (B) Luciferase activities in cells transfected with the truncated reporter constructs and pRL-TK. The transfected cells were cultured for 24 h in the presence of 10 mM IPTG, and then cell lysates were assayed for firefly and Renilla luciferase activities. Protein concentrations of cell lysates were determined by the Bradford method using bovine serum albumin as the standard protein, and firefly luciferase activity was normalized to light units (LU) per microgram of protein. Hatched bars indicate the promoter activity in U937/NS13 cells, and white bars indicate that in U937/CAT12 cells. Firefly luciferase activity in U937/NS13 cells transfected with the positive-control construct pGL3-control was 902.2 ± 105.0 LU/μg of protein, and that in U937/CAT12 cells was 850.2 ± 185.9 LU/μg of protein, whereas the activity from the negative-control construct pGL-Basic in U937/NS13 cells was 0.5 ± 1.3 LU/μg of protein and that in U937/CAT12 cells was 0.4 ± 1.3 LU/μg of protein. Numbers in parentheses besides the pairs of bars indicate the increased induction (fold). Transfection efficiency, assessed by Renilla luciferase activity from pRL-TK, was similar in each assay. The means (± standard deviations [error bars]) of three separate experiments are shown.