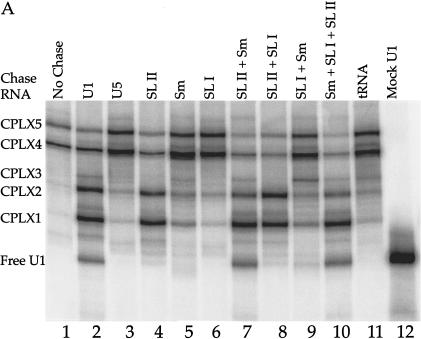
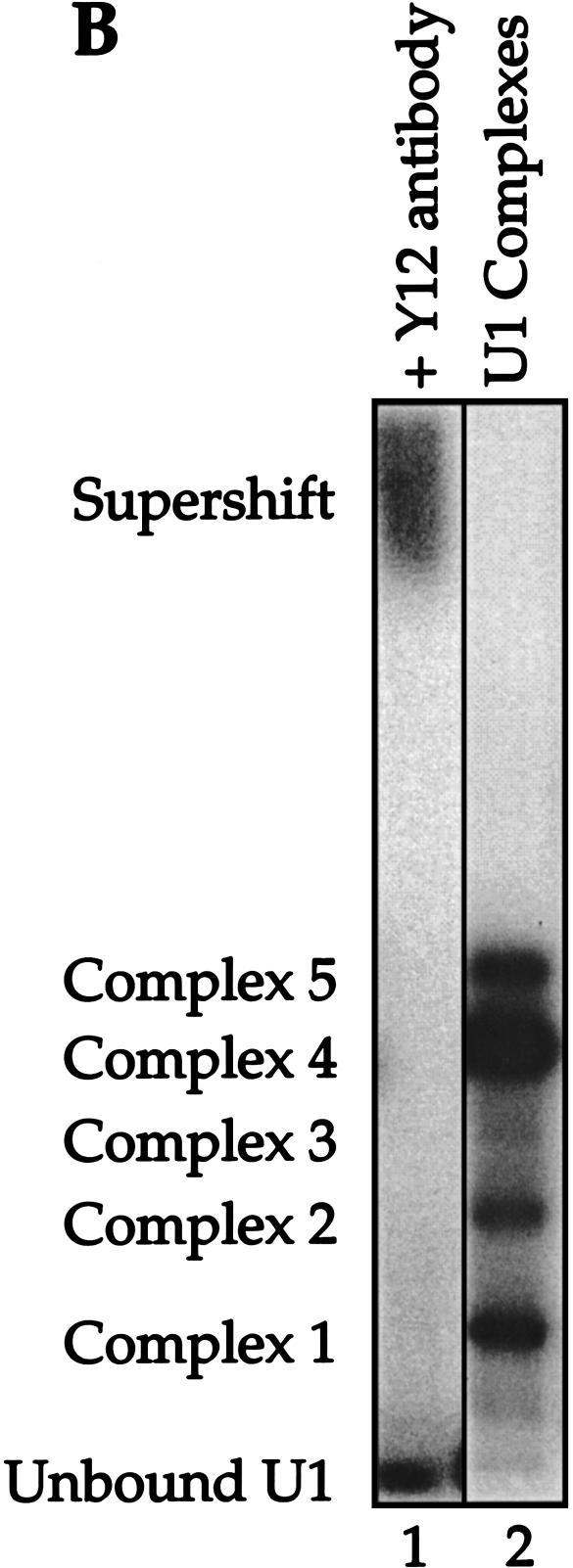
FIGURE 2.
Reconstituted U1 snRNPs dissociate into distinct faster-mobility species when challenged with excess cold U1 snRNA or other RNA fragments. (A) 32P-labeled U1 snRNA (10 nM) assembled with the core protein preparation to form two partial U1 snRNP species (lane 1). These U1 subparticles were then challenged with excess 4.6 μM unlabeled U1 snRNA (lane 2), 4.6 μM U5 snRNA (lane 3), 4.6 μM U1A stem loop II fragment, binding U1A (lane 4), 4.6 μM U1 Sm site fragment (lane 5), 4.6 μM U1 70K stem loop I fragment, binding U170K protein (lane 6), 4.6 μM each of pairs of U1 RNA fragments (lanes 7–9), or all three fragments (lane 10). Lane 11: 4.6 μM tRNA was also used as chase RNA. The U1 RNA fragments are described in Materials and Methods. Mock-treated 32P -labeled U1 RNA in the absence of protein is shown in lane 12. CPLX = complex. (B) The mobility of complexes 1–5 was retarded by the addition of Y12, an Sm protein-specific antibody.