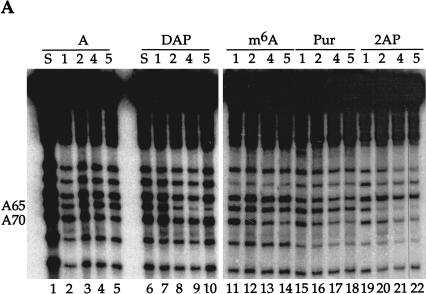
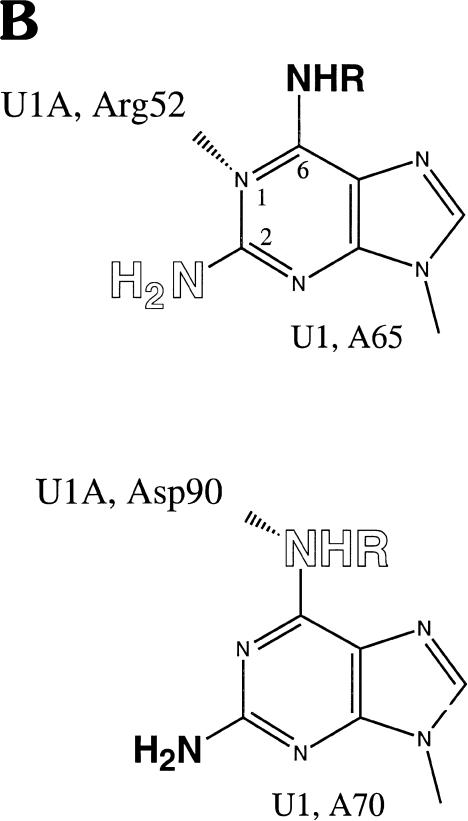
FIGURE 3.
U1A protein interactions with U1 snRNA defined by NAIM. (A) The sequencing gel shows iodine cleavage of RNAs isolated from native gel bands like those in Figure 2 ▶. The phosphorothioate-tagged analog incorporated into each U1 snRNA is noted above each set of lanes: DAP, 2,6-diaminopurine; m6A, N-methyl adenosine; Pur, purine; 2AP, 2-aminopurine. The numbers immediately below each nucleotide indicate the complex (1, 2, 4, or 5) from which the RNA was extracted. The A lanes (1–5) represent the control phosphorothioate isolated from each complex, and the S lanes are sample control lanes demonstrating phosphorothioate incorporation. For recognition by the U1A protein, an amino group at the 6-position is required on A70, but not on A65; an amino group at the 2-position is permitted on A70, but not on A65. (B) Proposed interactions of A65 and A70 with U1A residues. The U1A residue (Arg 52 or Asp 90) is shown interacting with a preferred position on the adenosine base (N1 of purine 65 or the exocyclic amine at the 6-position of purine 70). Allowed modifications are shown in bold, and those that interfere are in open letters. The NHR representation of the amine at the 6-position indicates that the absence of this amine or its modification (when R is a methyl group) is tolerated; the NH2 at the 2-position indicates that the presence of an amine is tolerated.