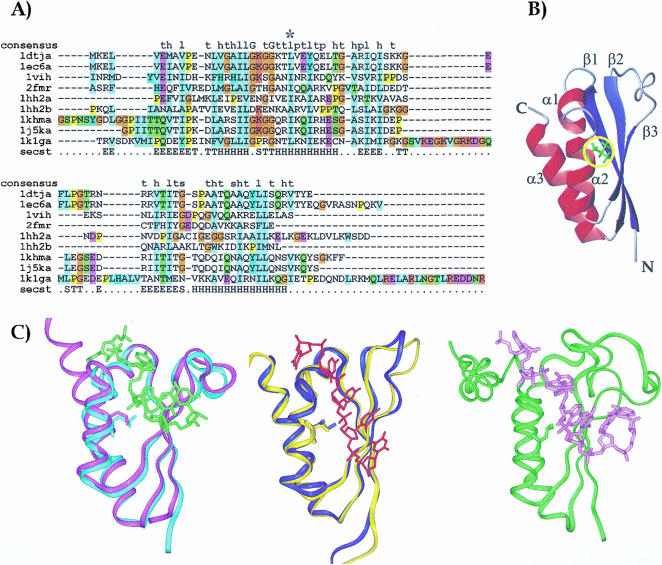
FIGURE 1.
(A) Sequence alignment of the KH constructs whose structure is currently known. Sequence alignment was obtained by the Clustalx program (Thompson et al. 1997). The sequences are labeled with the names of the corresponding PDB entries: 1dtj and 1ec6 correspond to the isolated and the complexed Nova KH3 constructs respectively (Lewis et al. 1999, 2000); 1vih, 2fmr, 1hh2-1 and 1hh2-2 are the sequences of vigilin KH6 (Musco et al. 1996), FMRP KH1 (Musco et al. 1997), and of the two KH repeats of NusA (Worbs et al. 2001); 1khm and 1j5k span the sequences of the isolated and complexed hnRNP K KH3, respectively (Baber et al. 1999; Braddock et al. 2002a); 1k1g is the sequence of the complexed Sf1 (Liu et al. 2001). A star indicates position α2(3). The consensus sequence as given by SMART is indicated in the first row (Consensus). The secondary structure of Nova is indicated in the last row (Secst). The DSSP convention was used (Kabsch and Sander 1983). (B) A ribbon representation of the KH fold. The coordinates of Nova KH3 have been used (Lewis et al. 1999). Red ribbons and blue arrows indicate the α helices and the β strands, respectively. The side chain of position α2(3) is shown in green and underlined by a yellow circle. (C) Structure comparison of KH structures. The protein backbones are displayed as a ribbon, and the α2(3) conserved amino acid is displayed in full. The structures were superposed by the Dali server (Holm and Sander 1993) and displayed by InsightII (Accelerys). (Left) Superposition of the free (cyan ribbon) and RNA complex (magenta ribbon with the RNA indicated in green) structures of Nova-2 KH3 (Lewis et al. 1999, 2000). (Middle) Superposition of the free (yellow ribbon) and RNA complex (blue ribbon with the RNA indicated in red) structures of the KH3 of hnRNP-K protein (Baber et al. 1999; Braddock et al. 2002a). (Right) KH-QUA2 structure in complex with RNA (green ribbon with the RNA in pink; Liu et al. 2001).