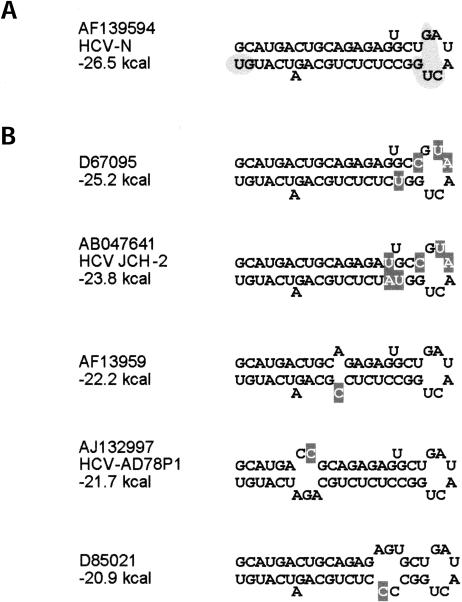
FIGURE 10.
Natural genetic variation within the SL1 sequence. (A) MFOLD-predicted structure of the SL1 sequence present in Ntat2ANeo(SI). Shaded regions indicate regions of critical sequences/structure as determined in this study. A BLASTN search of this 46-nt sequence revealed 79 hits in GenBank that were identified as HCV specific. Of these, 41 sequences are identical to the sequence shown in this panel. (B) Variant SL1 sequences found in the BLASTN search. Shown are those sequences in the database with multiple nucleotide substitutions, or those with single nucleotide substitutions that are present in more than one sequence in the database. Differences from the NtatNeo2A(SI) sequence are highlighted. The structures shown were predicted by MFOLD. GenBank accession numbers are shown for each example.