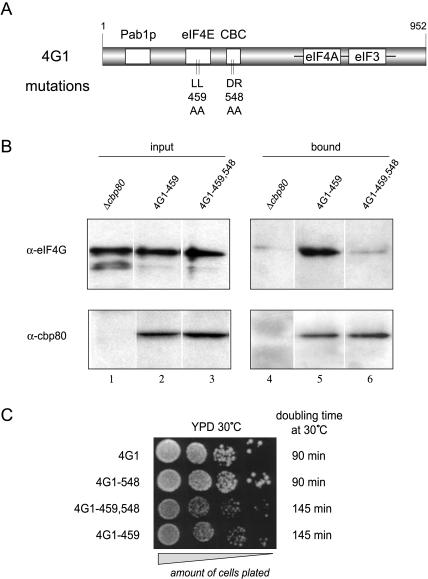
FIGURE 2.
Phenotypic characterization of the eIF4G1–548 mutation. (A) Schematic representation of the eIF4G1 protein. Boxed regions represent binding sites for the proteins indicated above. Vertical bars indicate the nature and position of the mutations described in this study. (B) Immunoprecipitation using rabbit polyclonal α-CBP80 antibodies and extracts from indicated yeast strains. A Western blot analysis is shown revealing the amount of eIF4G (upper panel) and CBP80 (lower panel) proteins present in the input (left panels, 1% of the input is loaded), and bound fractions (right panels, 100% of the eluate) of the immunoprecipitation. A residual level of eIF4G binding to the antibody resin is seen even in the absence of CPB80 protein (lane 4). Variation of several parameters of the immunoprecipitation protocol failed to reduce this background binding (data not shown); however, robust binding of eIF4G1–459 protein and a lack of detectable binding of eIF4G1–459,548 protein to the CBC resin was consistently seen in three independent repeat experiments. (C) Serial dilutions (10-fold) of the indicated yeast strains growing exponentially were plated on YPD and incubated for 48 h at 30°C. Doubling times listed on the right were measured in liquid YPD medium over 8 h of incubation at exponential growth at 30°C.