FIGURE 5.
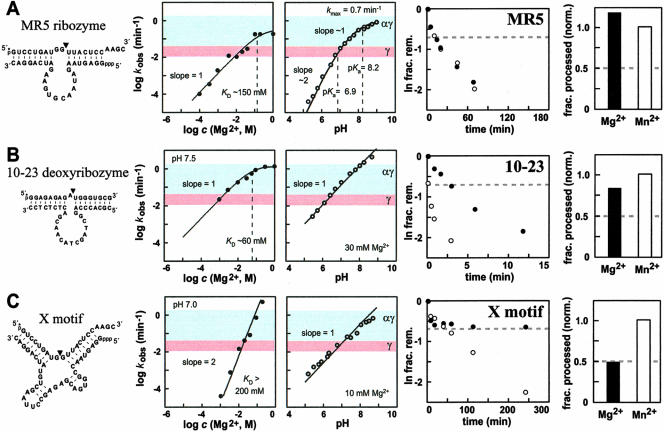
Enzymes with more complicated kinetic profiles can meet or break the αγ speed limit. (A–C) Sequences, secondary-structure models, and characteristics for the MR5, 10–23, and X-motif enzymes, respectively. The cofactor and pH dependencies for each enzyme are depicted in the plots to the left, and the data resulting from thio-effect examination are given in the plots to the right. Other notations are as described in the legends to Figures 2 ▶–4 ▶. Reagent concentrations (enzyme, KCl, and cofactor, respectively) were as follows for each construct: MR5: 24 nM, 250 mM, 200 mM; 10–23: 100 nM, 0 mM, 30 mM; X-motif: 50 nM, 0 mM, 20 mM. Below pH 7, assays of MR5 were buffered with Bis-Tris. Assays of X-motif were buffered with HEPES in the range pH 6.7 to pH 8. Portions of the data presented in Figure 5C ▶ were obtained from Lazarev et al. (2003) for comparison.