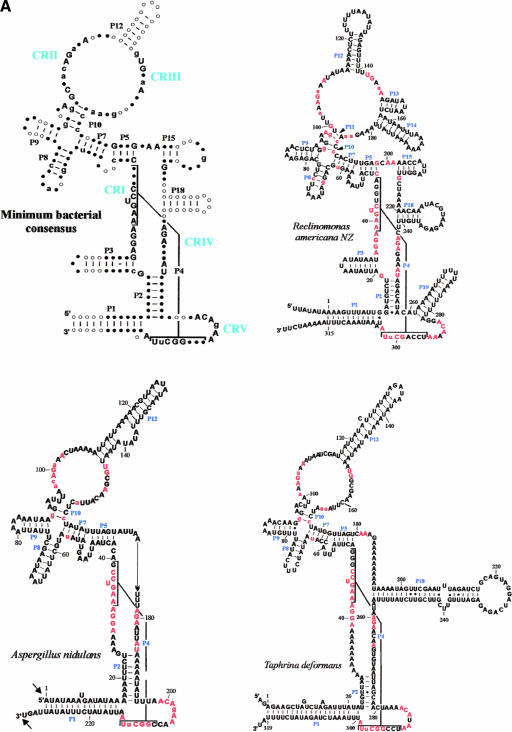
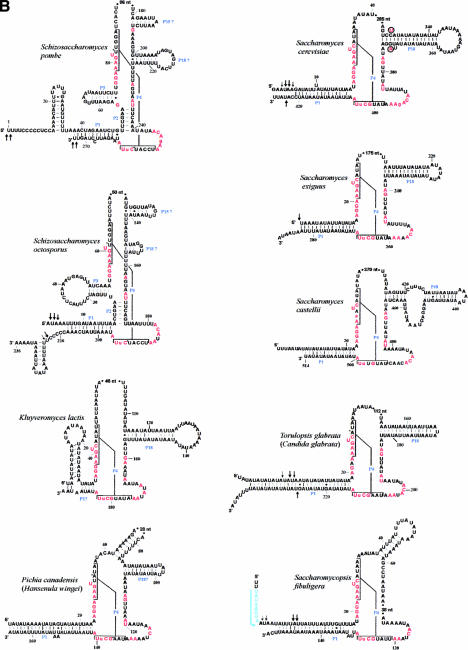
FIGURE 2.
Minimum bacterial P-RNA consensus and mtP-RNA secondary structure models of R. americana and ascomycete fungi. (A) Positions in red are invariant in the minimum bacterial consensus; uppercase letters in the mtP-RNAs indicate 100%, lowercase letters at least 90%, conservation of the minimum bacterial consensus sequence. The conserved motifs CRI through CRV are indicated in blue. The arrows indicate experimentally determined RNA extremities; arrow thickness is proportional to the percentage of molecules ending at this position. (B) mtP-RNA secondary structures from nine additional ascomycetes. In the S. cerevisiae structure, nucleotides that co-vary with the Saccharomyces douglasii sequence are in red circled letters. A putative promoter in S. fibuligera is highlighted in blue. Asterisks next to nucleotides in the P4 of S. pombe, S. octosporus, C. glabrata, and P. canadensis indicate mispairs in these helices. Note that the Candida structure P1 helix can be further extended by 9 to 12 bp from its mature ends. It is possible that the noncanonical A–G pair adjacent to the RNA processing sites in this mtP-RNA serves as a signal for RNA maturation.