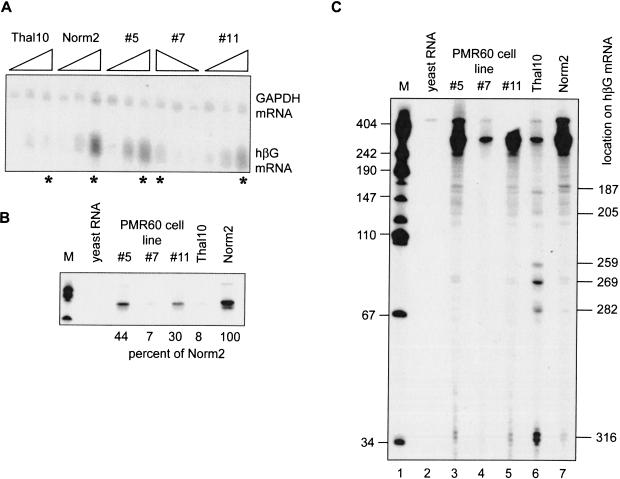
FIGURE 4.
Identification of β-globin mRNA decay intermediates in Norm2 cells expressing Xenopus PMR1. Cytoplasmic RNA was isolated at time 0, 48, and 96 h following DMSO-induction from Norm2, Thal10, or three lines of Norm2 cells stably transfected with plasmids expressing myc-PMR60. (A) Each of the RNA samples was analyzed by Northern blot that was hybridized to a mixed probe for β-globin and GAPDH mRNA. The RNAs were applied in the order indicated above with the exception of cell line #7, in which they were applied in reverse order. (B) Equal amounts of the 96-h samples identified with by an asterisk (*) in A were analyzed by S1 nuclease protection using a NaeI–BamH1 probe, and the protected products were analyzed as described previously (Stevens et al. 2002). An 18-h exposure of the portion of the gel corresponding to the fully protected probe is shown in B. The marker (M) in lane 1 consists of HpaII fragments of pUC19 DNA. (Lane 2) Yeast RNA was added to the initial hybridization reaction; (lanes 3–5) S1 nuclease protection was performed on RNA obtained from three independent clones of Norm2 cells stably transfected with myc-PMR60; (lanes 6,7) S1 nuclease protection was performed on RNAs from Thal10 and parental Norm2 cells. The relative quantity of each fully protected product was determined by scanning densitometry, and the percent of the signal for β-globin mRNA from Norm2 cells is shown beneath the gel. (C) The gel was exposed for 96 h to visualize β-globin mRNA decay intermediates. Human β-globin mRNA decay products characterized previously (Stevens et al. 2002) are identified on the right side of the autoradiogram.