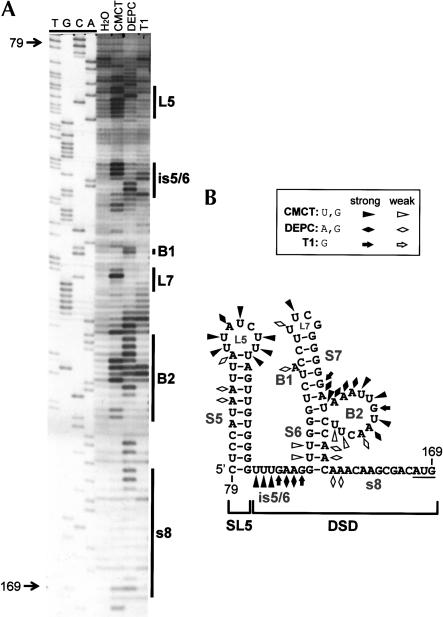
FIGURE 2.
(A) Chemical and enzymatic probing of the 3′ half of the TBSV 5′ UTR. In vitro transcripts of full-length DI-72SXP DI RNA were treated with CMCT [1-cyclohexyl-3-(2-morpholinoethyl)carbodiimide], DEPC (diethylpyrocarbonate), or RNase T1, and the cleaved or modified RNAs were analyzed by primer extension (PE). The PE products were separated in an 8% acrylamide-urea gel along with a control H2O-treated sample (H2O) and a sequencing ladder. Nucleotide positions relative to the TBSV genome are indicated on the left, whereas the locations of various single-stranded RNA elements are delineated by vertical lines on the right. (B) RNA secondary structure model for the 3′ half of the 5′ UTR. Chemical and enzymatic modifications have been mapped onto the MFOLD-predicted structure and are labeled according to the figure legend. Coordinates are given at the beginning and end of the sequence, and stem–loop (SL) and bulged (B) structures are labeled.