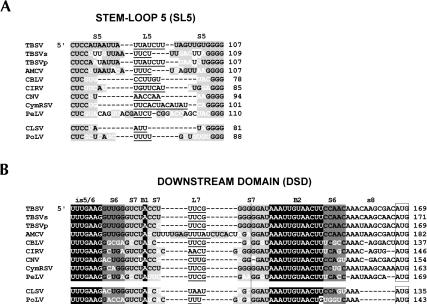
FIGURE 3.
Comparative sequence analysis of (A) SL5 and (B) DSD from members of the genera Tombusvirus and Aureusvirus. The tombusviruses are Tomato bushy stunt virus-statice isolate (TBSVs), Tomato bushy stunt virus-pepper isolate (TBSVp), Artichoke mottled crinkle virus (ACMV), Cucumber Bulgarian latent virus (CBLV), Carnation Italian ringspot virus (CIRV), Cymbidium ringspot virus (CymRSV), Cucumber necrosis virus (CNV), and Pear latent virus (PeLV). The aureusviruses are Cucumber leaf spot virus (CLSV) and Pothos latent virus (PoLV). Gaps (dashes) were introduced to maximize alignment. Subelements (i.e., stems, loops, and bulges) within the RNA structures are labeled above the sequences. Predicted single-stranded residues are depicted in black text (no highlighting) with loop regions underlined. Base-paired regions are in black text with light or dark gray highlighting. Residue differences in base-paired regions that maintain base-pairing via either mono- or covariation are indicated in white. Conserved single-stranded sequences in the DSD are depicted in white with black highlighting. In B the vertical line delineates the upper (on left) and lower (on right) portions of S7, and the open boxes enclose start codons for p33/92 (tombusviruses) and p25/84 (aureusviruses). The numbers on the right correspond to coordinates for the 3′-most residues shown.