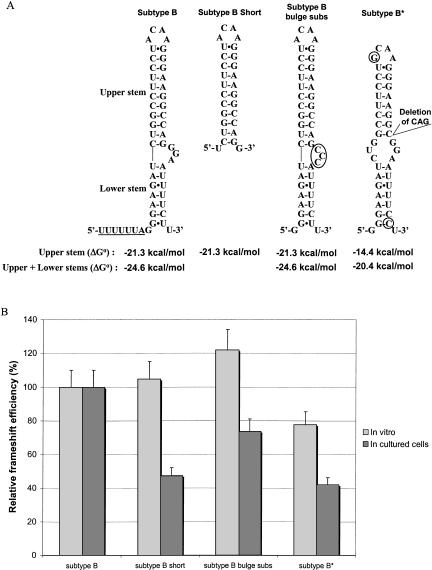
FIGURE 2.
Relative frameshift efficiency of mutants of subtype B of HIV-1 group M. (A) Secondary structure of the frameshift stimulatory signal for subtype B, subtype B short, subtype B bulge subs, and subtype B*, a natural variant that has a deletion of three nucleotides in the frameshift stimulatory signal. The stimulatory signal is represented with the extended bulged stem–loop structure found for HIV-1 subtype B (Dulude et al. 2002). Circles indicate nucleotides that differ from subtype B. The thermodynamic stability, ΔG° (kcal/mole), of the frameshift stimulatory signal of the different mutants of subtype B is indicated. The predicted ΔG° were calculated with the mfold program of M. Zuker (Mathews et al. 1999) for the classic stem–loop structure (upper stem) and the extended bulged stem–loop structure (upper stem + lower stem). The complete frameshift region with the slippery sequence underlined is represented for subtype B, whereas only the frameshift stimulatory signal is shown for the mutants. (B) Relative frameshift efficiency of mutants of subtype B. In vitro studies in RRL and assays in cultured cells (293T) were performed as described in the text. For assays in cultured cells, the experimental values were normalized for variations in transfection efficiency by cotransfecting the various constructs used for measuring the frameshift efficiency with pcDNA3.1/Hygro(+)/lacZ and monitoring the β-galactosidase activity. The frameshift efficiency of subtype B is arbitrarily set at 100%. The values of the frameshift efficiency are the means of at least five independent experiments, with bars representing the standard error on the means.