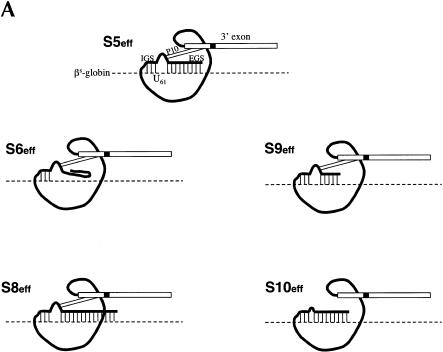
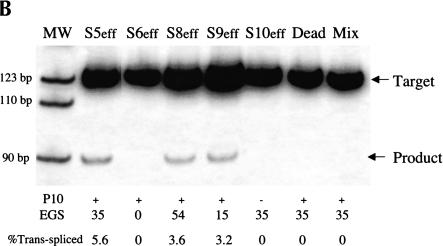
FIGURE 3.
Evaluating the importance of the P10 and EGS–target interactions for trans-splicing (A) Derivatives of the S5eff ribozyme with modified P10 or EGS sequences. Ribozyme derivatives pairing to the βS-globin target RNA (dashed line) are shown. The shaded box present in the ribozyme 3′ exon represents the priming site used to quantitate trans-splicing efficiency by QC RT–PCR. Compared to the 35-bp interaction between the external guide sequence (EGS) of the S5eff ribozyme and the target RNA, S6eff, S9eff, and S8eff ribozymes include an EGS that can form 0 bp, 15 bp, and 54 bp, respectively. Although the EGS is unmodified in S10eff, the P10 helix is disrupted. (B) Trans-splicing requires P10 and EGS pairing interactions. Expression constructs encoding the ribozymes described in (A) were co-transfected in 20-fold molar excess with βS-globin plasmid. The percentage of trans-spliced products were determined as described in Figure 2 ▶. For each ribozyme construct, the presence (+) or absence (−) of P10, and the length of the EGS interaction in base-pairs is indicated. Control transfections (“Dead” and “Mix”) were performed as in Figure 2B ▶. Molecular mass markers (MW) are shown.