FIGURE 4.
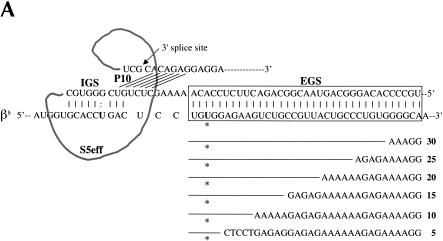
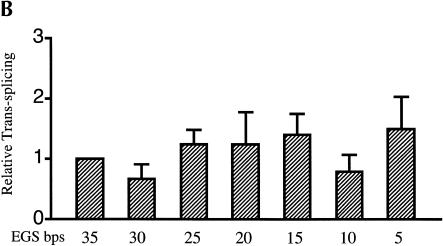
Evaluating EGS–target RNA base-pairing requirements. (A) Modifications to the βS-globin target sequence. The interaction between the S5eff ribozyme and the βS-globin target RNA is shown in detail. The EGS–target interaction is highlighted in the boxed region. Derivatives of βS-globin target RNA are represented below the boxed region. Modular mismatches of 5 bp were cumulatively introduced into the target to disrupt increasing regions of the EGS–target interaction. Unaltered regions of the target sequence are represented by the lines. The nucleotide sequences of the mismatched regions in the target RNA are shown. The number of base-pairs remaining in the EGS–target interaction is indicated in bold type. The sickle mutation is denoted with an asterisk. (B) Relative trans-splicing efficiency of the S5eff ribozyme with target RNAs containing mismatches in the region complementary to the ribozyme EGS. The matched (35 EGS bp) and mismatched (5–30 EGS bp) target RNAs were co-expressed with the S5eff ribozyme in HEK293 cells. Trans-splicing efficiency was quantitated by QC RT–PCR analysis for multiple independent transfection experiments (n = 6). Efficiency of trans-splicing was compared to the efficiency of S5eff trans-splicing with the matched βS-globin target.