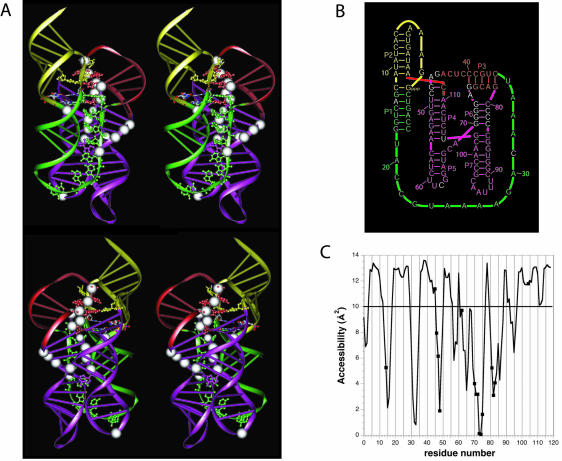
FIGURE 3.
A three-dimensional model for the class I ligase. (A) Two stereo views of the model. White spheres indicate those residues protected from hydroxyl-radical cleavage. Residues of particular interest are highlighted with all-atom representation. These include the residues of the proposed tandom G•A pairs and residues that both were unpaired in previous representations of the ligase secondary structure (Fig. 1A ▶) and were invariant among 25 active variants of the ligase (Ekland and Bartel 1995), all colored the same as the proximal paired regions. Also shown in all-atom representation are the four residues comprising the 2 bp flanking the ligation junction, colored according to the identity of the atoms. (B) A revised secondary structure diagram of the ligase that better reflects the arrangement of helices proposed by the tertiary model. Watson–Crick pairing is the same as in the original secondary structure (Fig. 1A ▶), except for one additional base pair, G47:C111, near the catalytic site. The color scheme reflects that of the ribbon representations in panel A. (C) Solvent accessibilities of the C4′ atoms in the modeled structure, as calculated using NACCES and a 1.4 Å sphere radius with an averaging over a window of three residues. A cut-off of 10 Å2 between accessibility and nonaccessibility is indicated by a horizontal line.