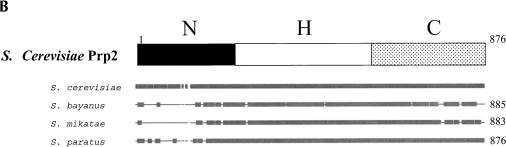
FIGURE 4.
Conservation of Prp2. (A) Alignment of the C-terminal region of Prp2 with other DExD/H family members shows conservation surrounding the D845N/C846Y mutations. Amino acids 780–876 of Prp2 were aligned with Prp16, Prp22, and Prp43 using the BLASTP program. Amino acid identity with Prp2 is shown as a black background with white letters, and conserved residues are shown in a gray background with white letters. The site from which a deletion is known to be lethal is indicated above the sequence by >; #, deletion from here has a temperature-sensitive growth phenotype, but is lethal in combination with a shortened N terminus; *, deletion from here has a temperature-sensitive growth phenotype; ~, deletion has no effect on growth; STOP, stop codon; numbers in parentheses indicate the number of amino acids to the end of the protein. (B) Comparison of the Prp2 protein from S. cerevisiae to the ortholog from three closely related Saccharomyces species, S. bayanus, S. mikatae, and S. paratus. The alignment was performed using Align Plus 4, Version 4.1 (Scientific & Educational Software) with a similarity significance cutoff equal to or greater than 90%. Areas of gray represent windows of 10 amino acids in length where the protein is 90% identical to the cerevisiae Prp2. S. bayanus Prp2 is 81% matched with S. cerevisiae, S. mikatae is 82% matched, and S. paratus is 90% matched. Lines in the diagram indicate less than 90% identity; gaps indicate deletions in the alignment. A representative version of the domain boundaries of Prp2 indicating N (N-terminal domain), H (Helicase domain), and C (C-terminal domain) relative to the alignment is provided on the top.