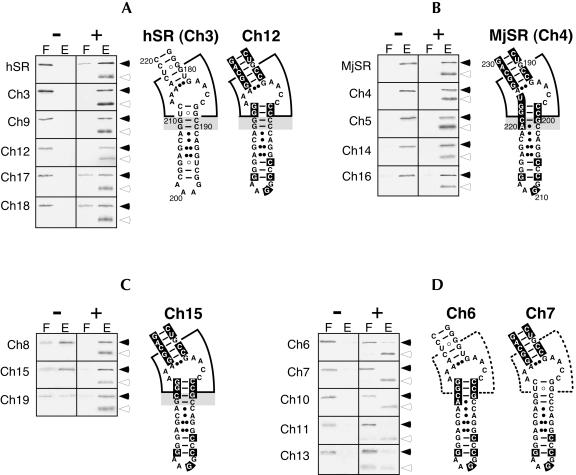
FIGURE 2.
SDS PAGE of human SRP54M (solid arrowhead) in the flowthrough (F) and high-salt eluate (E) of DEAE-columns in the absence (−) or presence (+) of SRP19 (open arrowhead). The RNAs used in the binding reactions are labeled on the left of each panel. (A) Mutant RNAs with assembly properties similar to those observed with human SRP RNA (hSR). (B) Mutant RNAs that behave like M. jannaschii SRP RNA (MjSR). (C) Functional mutant RNAs that exhibit a residual dependency on SRP19. (D) Mutant RNAs in which binding of SRP54M is significantly reduced or abolished. Secondary structure examples are given for hSR, MjSR, and mutant RNAs Ch3, Ch4, Ch6, Ch7, Ch12, and Ch15. The asymmetric loop and its surroundings are marked by a wedge, which is drawn with dashed line in D. (A–C) The 2 bp (C189–G210 and C190–G209 of hSR, or G200–C221 and C201–A220 of MjSR) crucial for SRP19 dependency are highlighted in gray.