FIGURE 2.
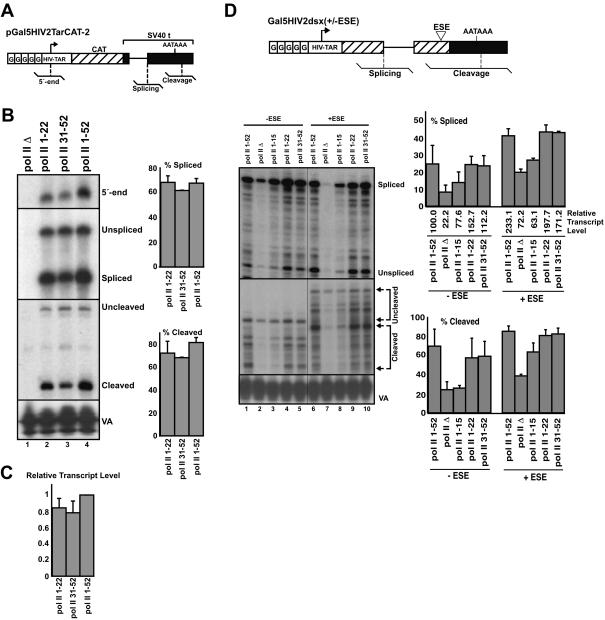
A CTD of 22 repeats is sufficient for efficient transcription, splicing, and 3′-end cleavage of constitutively spliced and ESE-dependent pre-mRNA reporter. (A) Schematic representation of the pGal5HIV2TarCAT-2 pre-mRNA reporter, which contains the HIV-2 promoter and five upstream Gal4 DNA binding sites, the chloramphenical acetyl transferase (CAT) gene, and the SV40 t intron and cleavage/polyadenylation signal. Transcriptional initiation, splicing, and 3′-end cleavage were monitored using the RNase protection probes shown schematically below the reporters. (B) Cells transfected with α-amanitin-resistant pol II deletion mutant expression plasmids, and an expression plasmid for the activator Gal4-VP16, were treated with α-amanitin, then retransfected with the pGal5HIV2TarCAT-2 pre-mRNA reporter and the pol III expression plasmid pSP-VA. RNA was recovered and analyzed by RNase protection using probes for measuring transcriptional initiation (5′-end), splicing, and cleavage levels. Levels of the VA RNA were also determined by RNase protection, allowing for normalization of signals due to variations in transfection efficiency and RNA recovery. Representative analyses from five independent repeat experiments are shown. Quantification of splicing and cleavage efficiencies is shown in the adjacent bar graphs. RNA signals were first corrected for the number of U-residues before calculating the percent spliced and cleaved transcripts. (C) Levels of transcripts shown in (B) were calculated by summing the U-corrected spliced and unspliced signals, and the U-corrected cleaved and uncleaved level signals, and dividing these values by the corresponding VA signals. The resulting values are represented as a fraction of the transcript level resulting from pol II 1-52. (D) Schematic representation of the Gal5HIV2dsx(+/−ESE) reporter, with RNase protection probes for monitoring splicing and cleavage indicated. The (+ESE) version of the reporter contains a synthetic (6xGAA repeat) ESE sequence within the 3′-exon; this ESE stimulates the recognition of the adjacent, suboptimal 3′ splice site. Cells were transfected with the α-amanitin-resistant pol II deletion mutant expression plasmids, activator Gal4-VP16, Gal5HIV2dsx either with (+ESE) or without (−ESE) the ESE, and pSP-VA. Cells were treated with α-amanitin, and recovered RNA was analyzed using the splicing and cleavage probes indicated in the diagram. RNA transcribed by pol II Δ from the Gal5HIV2dsx(+/−ESE) reporter was detectable above background levels, and therefore quantified. An α-amanitin-resistant pol II containing the first 15 CTD heptapeptide repeats, which expresses to comparable levels as pol II with a wild-type CTD (Fong and Bentley 2001), was also included for comparison. The bar graphs indicate quantification of splicing and 3′-cleavage levels from three repeat separate analyses, and were calculated as described in Fig. 1B ▶. Relative transcript levels were calculated as in Fig. 1C ▶, and are adjusted relative to the level for pol II 1-52 (−ESE reporter), which was set to 100.