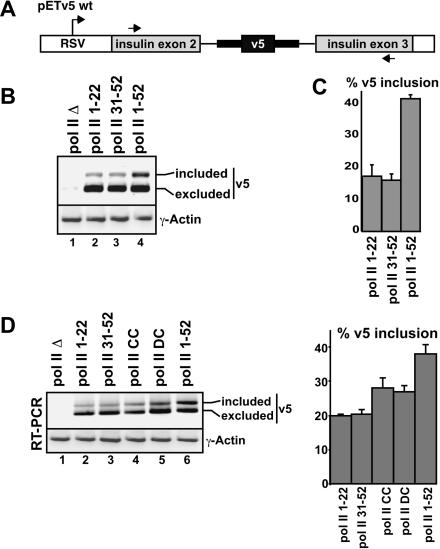
FIGURE 3.
Truncation of the CTD results in reduced inclusion levels of the alternatively spliced CD44 v5 exon. (A) Diagram of the pETv5 wt reporter minigene used to monitor alternative splicing. The variant v5 exon, from the human CD44 gene, is flanked by native CD44 intron sequences (indicated by a bold line) and intron/exon sequences from the constitutively spliced exon 2-intron 2-exon 3 region of the human insulin gene. Inclusion of v5 was detected by RT-PCR using primers specific for the constitutively spliced insulin exons, as indicated (Konig et al. 1998). (B) Human 293 cells were transfected with expression plasmids for the pol II deletion mutants indicated (refer to Fig. 1A ▶ ) and with pETv5 wt. Recovered RNAs were analyzed by RT-PCR. Spliced RNAs containing (included) or lacking v5 (excluded) are indicated. The level of γ-actin RNA was determined by RT-PCR as a control for RNA recovery. (C) The percent v5 inclusion measured from three independent experiments, with the average level and standard deviations shown. For quantification of the percent v5 inclusion, RT-PCR was performed using a radiolabeled PCR primer, as described in Materials and Methods, and signals were quantified by phosphorimager analysis. Linearity of signal detection was confirmed by comparing three different amounts of input RNA in the RT-PCR assays (data not shown). (D) RT-PCR analysis and quantification of the percent v5 inclusion levels in transcripts produced using additional pol II deletion mutants, as indicated. The experiment and analysis were performed as described in B and C.