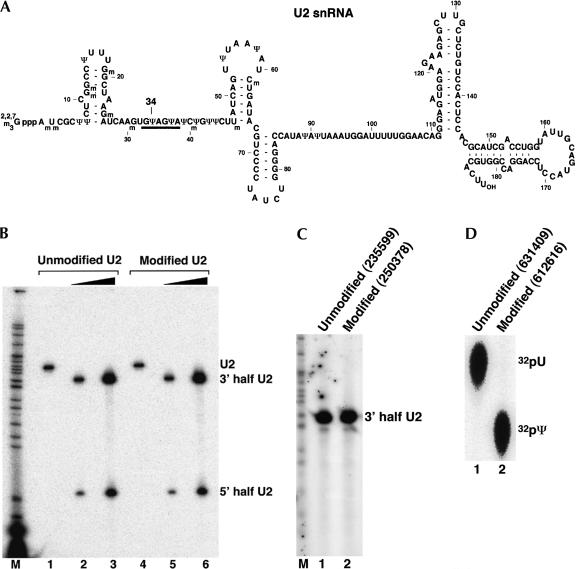
FIGURE 2.
Quantitative analysis of U2 snRNA pseudouridylation. (A) The sequence of mammalian U2 snRNA. Position 34 (to be analyzed) is indicated. The thick line denotes the bases involved in base-pairing interactions with the branch site sequence in pre-mRNA. (B) Site-specific RNase H cleavage directed by a 2′-O-methyl RNA–DNA chimera. Lightly radiolabeled unmodified U2 snRNA transcript (unmodified U2; lanes 1–3) and fully pseudouridine-substituted U2 transcript (modified U2; lanes 4–6) were subjected to RNase H cleavage in the presence of a 2′-O-methyl RNA–DNA chimera complementary to nucleotides 19–35 of U2 snRNA. The four 2′-deoxynucleotides in the chimera were designed to hybridize with nucleotides 30–33 of U2, leading to RNase H cleavage of the phosphodiester bond between positions 33 and 34 (target nucleotide). Reactions in lanes 2 and 4 contained 10 ng (0.15 pmole) of U2. Reactions in lanes 3 and 6 contained 70 ng (1.05 pmole) of U2. An overnight PhosphorImager exposure is shown, and the intact and cleaved U2 RNAs are indicated. (C) Replacement of the 5′-terminal phosphate with a radiolabeled phosphate (32P). The 3′-half RNA was treated with CIP and rephosphorylated with PNK and [γ-32P]ATP. The 3′-half RNA in lane 1 was derived from unmodified U2 and the RNA in lane 2 was from fully pseudouridine-substituted U2 (see B). A 1-min PhosphorImager exposure is shown. The numbers in parentheses are original PhosphorImager measurements (in volume). (D) Comparison of the radiolabeled nucleotides released from RNA. A small fraction of the 5′ endradiolabeled RNA derived from unmodified U2 (lane 1) or fully pseudouridine-substituted U2 (lane 2) was completely digested with nuclease P1. The released nucleotides (including the radiolabeled 5′-terminal nucleotide) were subsequently analyzed by TLC. The original numbers of PhosphorImager measurements are shown in parentheses. The positions of uridylate and pseudouridylate are indicated.