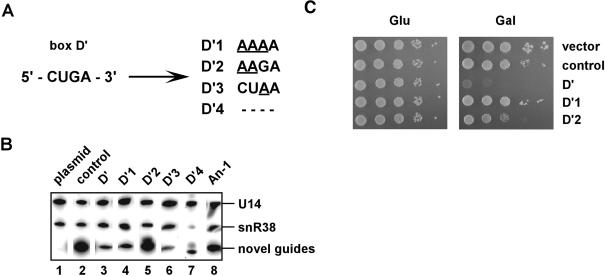
FIGURE 4.
Mutations in snoRNA elements required for methylation block the snoRNA interference effect. (A) Sites of mutation introduced into the novel U1757 (U1495) snoRNA. The wild-type box D′ motif (left) is shown, and mutations in this element are underlined (right). Deletion of box D′ is indicated with dashes. The set of mutant snoRNAs also included a variant with a point mutation in the antisense guide sequence, located 4 nt upstream of box D′ (An-1, data not shown). (B) Steady-state levels of the mutant snoRNAs. Northern blots were carried out on total RNA isolated from cells containing the following plasmids: empty plasmid (lane 1, plasmid); the parental control snoRNA (snR38), which was plasmid encoded (lane 2, control); the initial U1757 (U1495) guide snoRNA (lane 3, D′); and the experimental mutant variants of the U1757 (U1495) snoRNA (lanes 4–8). Natural U14 and snR38 snoRNAs derived from the genome served as internal controls. RNAs were separated on a urea-8% polyacrylamide gel, and blots were probed with a mixture of 5′-labeled oligonucleotides specific for snR38 and U14 snoRNAs. The snoRNA genes are identified at the top of the panel by the mutated elements. (C) Growth properties of cells with the mutant guide RNA genes. Transformants were diluted serially 1:10 and spotted on solid selective medium containing glucose (Glu) or galactose (Gal).