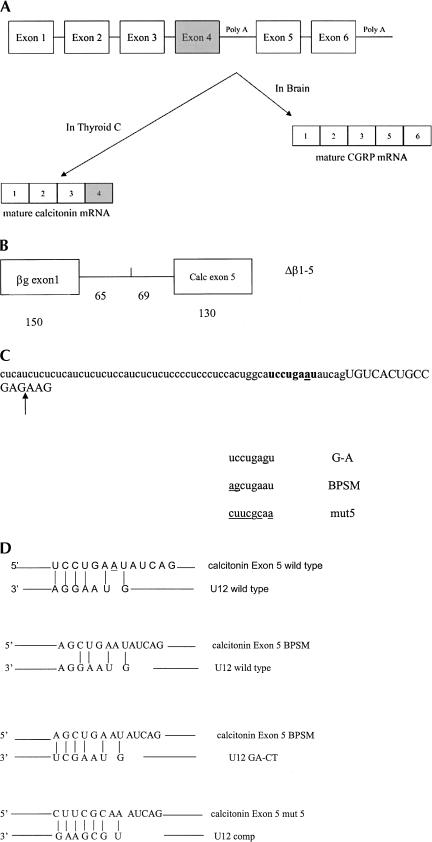
FIGURE 1.
The rat calcitonin/CGRP gene. (A) Structure and alternative RNA processing of the rat calcitonin/CGRP gene, exon sequences are denoted by boxes. (B) The Δβ 1-5 minigene structure showing the sizes in nucleotides of the exon and intron segments. (C) Sequence of the rat calcitonin/CGRP gene at the exon 5 splice acceptor. Intronic sequence is in lowercase, and exon 5 sequence is in capitals. A candidate U12 branch sequence is in bold, and the position of cryptic splicing is indicated by the arrow. The sequence of the A-G, BPSM and mut5 base changes in the putative U12 branch sequence are also shown, with base changes underlined. (D) Predicted interactions between the candidate branch sequence upstream of exon 5 and U12 snRNA. The first alignment is between wild-type calcitonin/CGRP and wild-type U12, the second contains the calcitonin/CGRP BPSM base changes, the third calcitonin/CGRP BPSM with U12 containing compensatory base changes, and the fourth potential base-pairing between calcitonin/CGRP mut5 and U12comp.