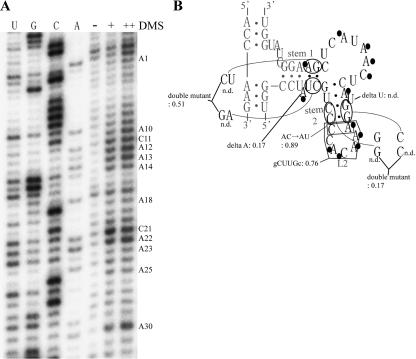
FIGURE 5.
Secondary structure analysis and site-directed mutations of the selected sequence in clone I RNA. (A) Autoradiogram of the DMS modification of clone I RNA. Modified bases are indicated at right. (B) Secondary structure model of clone I. Bases modified with DMS are indicated with black circles. The nucleotides derived from the randomized sequence or the constant region, are in black or gray, respectively. Relative activity is indicated for each mutant by using clone I as the standard (1.00). (n.d.) Nondetectable low activity. Conditions: 80 mM of MgCl2, 50 mM of KCl, 30 mM of Tris-HCl (pH 9.0), 0.5 μM of substrate RNA, and 50 nM of the ribozyme at 37°C for 1 h.