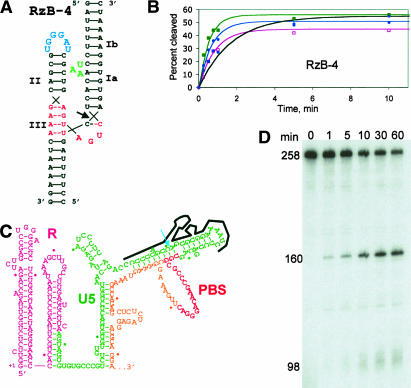
FIGURE 5.
Cleavage of HIV-1 RNA at submillimolar MgCl2. (A) Predicted structure of ribozyme RzB-4 annealed to a 23-nt substrate derived from the genomic RNA of HIV-1 strain HXB2. (B) Plots of substrate cleavage by RzB-4 under same conditions as in Figure 4. (C) Predicted secondary structure the HIV-1 genomic 5′ leader, based on comparison between SIV and HIV sequences (Rivzi and Panganiban 1993). Note that other secondary structures have also been proposed for this region. The first 242 nt of the 258-nt transcript are shown. Annealed RzB-4 is shown schematically, with cleavage site indicated by the arrow. (D) Cleavage reaction of internally radiolabeled, 258-nt synthetic genomic transcript in 0.5 mM MgCl2 at 37°C (pH 7.5), in the presence of a DNA oligonucleotide to mimic binding of the natural tRNA3Lys replication primer. Reaction times are given in minutes above the lanes. Cleavage products at 160 and 98 nt are indicated.