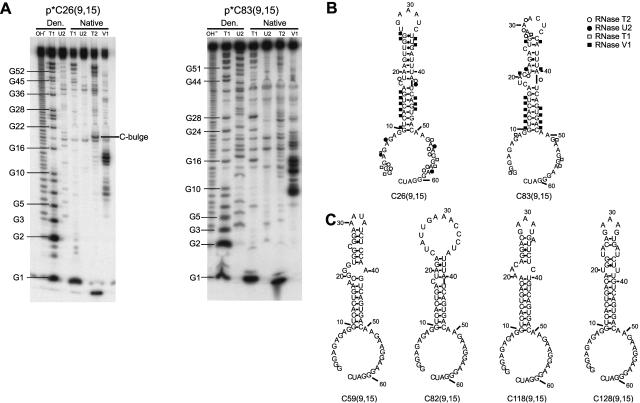
FIGURE 2.
Secondary structures of selected RNAs. (A) Structure mapping of two representative aptamers. Denaturing 12% polyacrylamide gels are shown. p*C26(9,15) is 5′-end labeled clone 26 with 5′- and 3′-tails of 9 and 15 nt, respectively, and is from the selection using K296R; p*C83(9,15) is 5′-end labeled clone 83 with the same tails and is from the selection using p20. Lanes are as follows: OH- is a limited alkaline digest; T1, U2, T2, and V1 are limited digests with ribonucleases specific for single-stranded G’s, A’s, N’s, or double-stranded regions, respectively. “Den.” denotes treatment with nucleases under denaturing conditions, while “Native” denotes treatment with nucleases under nondenaturing conditions. (B) Secondary structural models derived from structure mapping experiments in panel A. Positions of cleavage by single- and double-stranded probes are shown; symbols are provided in the figure. The five 3′-most nucleotides were present in the run-off transcripts from the final clones only. (C) Predicted secondary structures of representative selected RNAs determined using mfold v3.1 (Mathews et al. 1999; Zuker et al. 1999). Clones 59 and 82 are from the selection using p20, and clones 118 and 128 are from the selection using K296R.