Abstract
In addition to triggering nonsense-mediated mRNA decay (NMD), premature translation-termination codons (PTCs) frequently induce alternative splicing, an observation referred to as nonsense-associated alternative splicing (NAS). In many cases, NAS is induced because the nonsense mutation alters a splicing signal, such as inactivating an exonic splicing enhancer. However, for a few genes, NAS was reported to be PTC specific, implying that a translation signal could influence splicing. Here, we investigated whether production of a previously undetected alternatively spliced transcript from immunoglobulin μ (Ig-μ) depends on premature termination of the open reading frame. We show that PTCs at different positions in the VDJ exon of an Ig-μ minigene activate usage of an alternative 3′ splice site, generating an alternative transcript that lacks the initial PTC and a previously identified NMD-promoting element (NPE), but contains new PTCs because of a frame shift. Corroborating the importance of the NPE for maximal NMD response, the alternative transcript is only moderately down-regulated by NMD. We further demonstrate that NAS of Ig-μ minigene transcripts is not PTC specific. This finding, together with our results that contradict the previously reported frame dependence of TCR-β NAS, challenges the idea that cells might possess mechanisms that would allow regulation of splice site selection in response to premature termination of the ORF.
Keywords: premature translation-termination codon (PTC), nonsense-associated altered splicing (NAS), exonic splicing enhancer (ESE), immunoglobulin μ (Ig-μ)
INTRODUCTION
In addition to triggering nonsense-mediated mRNA decay (NMD) (Maquat 2004), premature translation-termination codons (PTCs) are also found to alter the splicing pattern in many cases, often leading to skipping of the PTC-containing exon (Li and Wilkinson 1998; Valentine 1998; Hentze and Kulozik 1999; Maquat 2001; Cartegni et al. 2002). The term “nonsense-associated altered splicing” (NAS) has been proposed to describe such changes in splicing caused by PTCs (Hentze and Kulozik 1999) and will be used herein.
Two conceptually different explanations for NAS have been proposed, and both are supported by respective experimental data. Many cases can be readily explained by conventional mechanisms involving chance disruption of conserved sequence motifs involved in regulating splice site choice, for example exonic splicing enhancers (ESEs) (Blencowe 2000; Graveley 2000). Accordingly, NAS was in many cases found to be a consequence of destruction of an ESE by the PTC-generating nonsense or frame-shift mutation (Valentine 1998; Liu et al. 2001; Caputi et al. 2002; Buchner et al. 2003; Pagani et al. 2003). In contrast, other studies provided evidence for PTC-specific NAS, indicating that premature termination of the translational reading frame affects splicing (Dietz and Kendzior 1994; Lozano et al. 1994; Aoufouchi et al. 1996; Gersappe and Pintel 1999; Mühlemann et al. 2001; Li et al. 2002; Wang et al. 2002c). The idea that the coding potential of an mRNA affects its own splicing is paradoxical and cannot be explained with our current mechanistic understanding of gene expression (for reviews, see Wilkinson and Shyu 2002; Dahlberg et al. 2003). Because of its far-reaching consequences for molecular models of gene expression, it is important to design experiments that allow unambiguous proof of the frame dependence of NAS, which is not trivial. In fact, only in a very few cases where NAS was observed has the issue of frame dependence been systematically addressed. So far, the most convincing evidence for frame-dependent NAS has been reported by Wilkinson and colleagues, who studied a mouse TCR-β minigene in human cells (Wang et al. 2002c). However, in contrast to their results, our own studies using some of these TCR-β minigenes did not reveal any correlation between production of the alternatively spliced mRNA and the truncation of the ORF (Mohn et al. 2005).
Because of the similarities between TCR-β and Ig-μ genes with regard to effects elicited by PTCs (Baumann et al. 1985; Carter et al. 1996; Mühlemann et al. 2001; Gudikote and Wilkinson 2002; Wang et al. 2002b; Bühler et al. 2004), we investigated whether PTCs in Ig-μ minigenes also induce NAS, and whether this NAS was PTC specific or not. We found that PTCs at different positions in the VDJ exon indeed lead to the production of a previously undetected alternatively spliced transcript by activation of an alternative 3′ splice site. This alternative Ig-μ transcript contains PTCs generated by a frame shift and is subjected to NMD. Importantly, we find no evidence for frame dependence of this Ig-μ NAS and therefore conclude that frame-dependent NAS, if existing at all, does not represent a general response to PTCs of transcripts encoded by genes of the immunoglobulin superfamily.
RESULTS AND DISCUSSION
PTCs located in the Ig-μ VDJ exon induce alternative splicing
The reports that PTCs located in the VDJ exon of a TCR-β transcript induce NAS that depends on truncation of the reading frame (Wang et al. 2002a,c) prompted us to investigate whether PTCs located in an Ig-μ VDJ exon also induce NAS, and whether this NAS was frame dependent. To address this, we used an Ig-μ minigene developed in our laboratory (Bühler et al. 2004) and first analyzed RNA from HeLa cells expressing either miniμ-wt, or miniμ constructs with a PTC at amino acid positions 57, 73, or 310 (Fig. 1A) by Northern blot analysis or conventional endpoint RT-PCR. Although Northern blot analysis revealed no major new Ig-μ species in cells expressing the PTC+ constructs, we observed in some experiments an additional very faint band migrating faster than the band corresponding to normally spliced Ig-μ mRNA when miniμ-ter73 was expressed, but not when miniμ-wt or miniμ-ter310 was expressed (Fig. 1B). A similarly smaller amplicon was also detected by RT-PCR with RNA from miniμ-ter57-expressing cells, but not when miniμ-wt or miniμ-ter310 was expressed (Fig. 1C). Cloning and sequencing of the smaller miniμ-ter57- and miniμ-ter73-specific RT-PCR products revealed an alternatively spliced transcript (hereafter called alt-mRNA), in which the 5′ splice site of the leader exon was spliced to an alternative 3′ splice site (alt-3′ ss) at amino acid position 96 in the VDJ exon. Usage of this alt-3′ ss results in skipping of the PTCs at amino acid positions 57 and 73, but induces a frame shift that generates new PTCs further downstream in this mRNA (Fig. 1A, out of frame). This is an important difference to other examples of NAS, where the ORF is restored in the alternatively spliced mRNA (Dietz and Kendzior 1994; Li et al. 2002; Wang et al. 2002c), and inconsistent with the idea of NAS having evolved to avoid production of PTC+ mRNAs (Li et al. 2002).
FIGURE 1.
Activation of an alternative 3′ splice site by PTCs at different positions in the Ig-μ VDJ exon. (A) Schematic representation of the Ig-μ gene with numbers depicting the amino acid positions where PTCs were inserted into the μ-minigene. The position of the PTC in the endogenous Ig-μ gene in two different hybridoma cell lines is indicated by the original clone name of the respective cell line (Connor et al. 1994). The PTC in N114 and U30 are at codons 73 and 335, respectively. Below, the alternatively spliced miniμ transcript is illustrated, in which an alternative 3′ splice site (alt-3′ ss) at amino acid position 96 in the VDJ exon is spliced to the 5′ splice site of the Vleader exon. Usage of the alt-3′ ss results in skipping of the PTCs, but induces a frame shift that generates new PTCs downstream in this mRNA (alt-mRNA, out of frame). The position of the single nucleotide insertion to restore the ORF in the alternatively spliced mRNA is depicted (−1FS; alt-mRNA, in frame). Exons are represented as boxes, introns as lines. The ORF of each transcript is indicated in white, untranslated regions in gray. The position of the TaqMan probe used to amplify alt-mRNA by real-time PCR is indicated as a black bar. (B) Northern blot analysis of RNA from polyclonal HeLa cell pools stably transfected with the indicated miniμ genes using a 32P-labeled probe to detect Ig-μ mRNA. In lane 4, miniμ-ter73-expressing HeLa cells were exposed to 100 μg/mL cycloheximide (CHX) for 2 h before analysis. As a loading control, the 28S rRNA band from the ethidium-bromide-stained gel before blotting is shown in the lower panel. (C) RNA of polyclonal HeLa cell pools stably transfected with the indicated miniμ genes were analyzed by RT-PCR. RNA from untransfected Hela cells serves as a negative control (HeLa). Cloning and sequencing of the specific RT-PCR product denoted as alt-mRNA (394 bp) revealed the alternatively spliced transcript (alt-mRNA) schematically represented in A.
Alt-mRNA levels are down-regulated by NMD
Because of the presence of PTCs in the Ig-μ alt-mRNA, we expected that these transcripts would be subjected to NMD. Indeed, when HeLa cells expressing miniμ-ter73 were treated with cycloheximide (CHX), a potent translation inhibitor that abrogates NMD (Carter et al. 1995), the level of alt-mRNA increased several-fold (Fig. 1B, lane 4). To increase alt-mRNA levels for easier detection, we introduced a −1 frame shift (−1FS) downstream of the alt-3′ ss into the constructs described above (Fig. 1A, in frame). This frame shift restores the ORF for alt-mRNA, but shifts the frame in the normally spliced mRNA so that the normally spliced mRNA acquires a PTC at amino acid position 108. Consistent with the expectation that this frame shift results in stable, PTC-free Ig-μ alt-mRNA, a significantly stronger signal for alt-mRNA was detected by end-point RT-PCR for miniμ-ter57−1FS than for miniμ-ter57 (Fig. 1C, cf. lanes 2 and 5). Collectively, these results demonstrate that the Ig-μ alt-mRNA contains a PTC, which renders it susceptible for NMD.
To accurately quantify Ig-μ alt-mRNA levels, we pre-formed real-time RT-PCR using a TaqMan probe that spans the alternative exon–exon junction and therewith allows specific detection of alt-mRNA without amplifying other potential splice variants (Fig. 1A). With this assay, we measured relative alt-mRNA levels in cells expressing miniμ-ter32, -ter57, -ter73, -ter310, and -wt (Fig. 2A, bars “out of frame”) and in cells expressing the corresponding constructs with the −1FS mutation (bars “in frame”). With the out-of-frame constructs, no alt-mRNA could be detected with miniμ-ter310 and only minute amounts with miniμ-wt, whereas alt-mRNA was readily detectable with the three constructs that had the PTC in the VDJ exon. Compared to the alt-mRNA level in miniμ-wt, alt-mRNA levels are 18-fold higher in miniμ-ter32, 15-fold higher in miniμ-ter73, and 80-fold higher in miniμ-ter57, respectively. The −1FS mutation increased alt-mRNA levels between 1.6- and 3-fold for miniμ-ter32, -ter57, and -ter73 (Fig. 2A, bars in frame), indicating that in the normal out-of-frame situation, alt-mRNA is down-regulated by NMD to this extent. Interestingly in the context of our recent identification of a NMD promoting element located in the 5′ half of the VDJ exon of the Ig-μ minigene (Bühler et al. 2004), this moderate effect of NMD on alt-mRNA observed here corroborates the importance of the 5′-half of the VDJ exon for efficient NMD, because alt-mRNA lacks this NMD-promoting sequence. In fact, the observed 1.6- to 3-fold difference between PTC+ (out of frame) and PTC- (in frame) alt-mRNA levels is similar to the threefold reduction of mRNA caused by the PTCs in constructs lacking the 5′-half of the VDJ exon (Bühler et al. 2004).
FIGURE 2.
(A) Alt-mRNA levels are down-regulated by NMD. Total cellular RNA of HeLa cells transiently transfected with the indicated miniμ genes were analyzed 48 h post-transfection by real-time RT-PCR. Ig-μ alt-mRNA was measured over the junction between the 5′ splice site of the leader exon and the alt-3′ splice site (see Fig. 1A), and relative Ig-μ alt-mRNA levels were normalized to neomycin mRNA encoded on the transfected plasmid. Average values and standard deviations of three real-time PCR runs of a typical experiment are shown. The alt-mRNA level measured for miniμ-ter57−1FS was set as 100%. White bars represent out of frame alt-mRNA levels, gray bars represent alt-mRNA levels with a restored open reading frame produced from constructs with a −1 frame shift (−1FS, see Fig. 1A). The signal obtained for miniμ-ter310 alt-mRNA was below the level of detection and set to zero. (B) Detection of the Ig-μ polypeptide encoded by the in-frame alt-mRNA by Western blot analysis. The antibody recognizing the constant region of the Ig-μ heavy chain detects a slightly faster migrating band for cells expressing miniμ −1FS constructs with a PTC in the VDJ exon. This band (Ig-μ alt) corresponds in size to the polypeptide encoded by the in-frame alt-mRNA, which is 80 amino acids (i.e., approximatively 9 kDa) smaller than the polypeptide encoded by the normally spliced mRNA, which is shown on the left as a marker (Ig-μ). Lamin C is shown as loading control. (C) A PTC in the VDJ exon of the endogenous Ig-μ gene also activates the alt-3′ splice site. RNA of the parental hybridoma cell line Sp6, encoding a PTC-free Ig-μ mRNA, and of three Sp6-derived, mutated cell clones was analyzed by real-time RT-PCR as in A. Relative Ig-μ alt-mRNA levels were normalized to 18S rRNA levels and average values and standard deviations of three real-time PCR runs of a typical experiment are shown. The alt-mRNA level of N114 was set to 100%. The PTC in N114 and U30 is located at amino acid positions 73 and 335, respectively (see Fig. 1A). X10 has a deletion of the whole Ig-μ gene and serves as negative control.
In-frame alt-mRNA is translated
Although we could detect and measure miniμ alt-mRNA easily with real-time RT-PCR, it is obvious from the Northern blot experiments (Fig. 1B; data not shown) that only a small fraction of the total miniμ pre-mRNA in a cell gets spliced by using the alt-3′ss. Therefore, we wanted to confirm the production of alt-mRNA by an independent method and decided to test whether the protein encoded by alt-mRNA expressed from -1FS constructs can be detected by Western blotting. As a reference and positive control, the Ig-μ polypeptide encoded by the normally spliced miniμ-wt mRNA is shown in Figure 2B, and miniμ-ter310 served as a negative control, because no alt-mRNA is produced from this construct. In agreement with the first 241 nt of the VDJ exon missing in alt-mRNA, an ∼ 9-kDa smaller band was detected in cells expressing −1FS miniμ constructs with PTCs in the VDJ exon (Fig. 2B). Notably, the amounts of μ polypeptide detected by Western blotting are roughly proportional to the alt-mRNA levels (Fig. 2, cf. A and B). This result shows that alt-mRNA, although much less abundant than the normally spliced mRNA, is translated into detectable amounts of protein when its open reading frame is restored.
A PTC in the VDJ exon of endogenous Ig-μ gene also activates the alt-3′ splice site
Next, we asked whether the alt-3′ splice site is also induced by a PTC in the endogenous Ig-μ gene expressed in mouse hybridoma cells. For this, we determined by real-time RT- PCR relative Ig-μ alt-mRNA levels in RNA isolated from the parental Sp6 cell line (wt Ig-μ), two cell lines expressing a PTC+ variant of the gene (N114 and U30; see Fig. 1A), and as a control from a cell line lacking the entire Ig-μ gene (X10). The result obtained from these endogenous Ig-μ genes (Fig. 2C) resembles the results with the Ig-μ mini-genes (Fig. 2A): Whereas only very small amounts of alt-mRNA could be detected in Sp6 and U30 (PTC at amino acid position 335 in exon C2), a PTC in the VDJ exon at amino acid position 73 (N114) elevated alt-mRNA >12-fold. Thus, the same NAS identified in HeLa cells expressing PTC+ miniμ genes also occurs in the endogenous Ig-μ gene, demonstrating that activation of this alt-3′ splice site by nonsense mutations is not cell type specific, nor an artifact of the Ig-μ minigenes.
Ig-μ alt-mRNA splicing is independent of hUPF1
Frame-dependent NAS would be expected to require “reading” of the ORF, i.e. translation or a translation-like scanning, and distinguishing between a premature and a physiological termination codon would be expected to rely on the same factors involved in PTC recognition during NMD. However, the data reported for TCR-β NAS is only partially consistent with these predictions. Whereas TCR-β NAS was inhibited by a stem-loop structure in the 5′ UTR, by expression of suppressor tRNA, or by removal of the initiator AUG codons, all indicating the requirement for frame-scanning, treatment of the cells with CHX did not abrogate production of alt-mRNA (Wang et al. 2002a,c). With regard to the requirement of NMD factors for NAS, depletion of hUPF2 did not inhibit alt-mRNA production (Mendell et al. 2002; Wang et al. 2002a), but depletion of hUPF1 did (Mendell et al. 2002).
Treatment of HeLa cells expressing miniμ-ter73 with CHX did also not inhibit production of alt-mRNA. In contrast, alt-mRNA levels increased, because NMD is inhibited by CHX, resulting in stabilization of the PTC-containing alt-mRNA (Fig. 1B, lane 4). This suggests that Ig-μ NAS differs from NMD in that it is not CHX-sensitive. To address whether Ig-μ NAS depends on hUPF1, we knocked down hUpf1 by RNAi in HeLa cells expressing miniμ-ter73. A substantial reduction of hUPF1 protein stabilized both the normally and alternatively spliced Ig-μ transcripts (Fig. 3). This result shows that hUpf1 knockdown inhibited NMD under our experimental conditions, but did not affect alternative splicing of Ig-μ pre-mRNA. Because no complete depletion of hUPF1 protein could be achieved, it is possible that Ig-μ NAS depends on hUPF1, and that the residual hUPF1 level after RNAi was not limiting for NAS. However, the result is also consistent with Ig-μ NAS being independent of hUPF1. Collectively, the increase of PTC+ alt-mRNA levels in response to CHX treatment (Fig. 1B) and hUPF1 knockdown (Fig. 3A) demonstrates that PTC+ alt-mRNA is a substrate for NMD and indicates that Ig-μ NAS acts upstream of NMD and independently of it.
FIGURE 3.
RNAi-mediated hUpf11 knockdown stabilizes miniμ-ter73 mRNA as well as alt-mRNA levels. (A) Northern blot analysis of total cellular RNA from HeLa cells expressing miniμ-ter73 using a 32P-labeled probe to detect Ig-μ mRNA. Knockdown of hUpf1 was induced by cotransfection of a GFP expression plasmid (pEGFP-N1) and a shRNA expression vector targeting hUpf1 mRNA (pSUPER-Upf1). RNA was isolated 72 h post-transfection from GFP positive cells sorted by FACS. As a loading control, the 28S rRNA band from the ethidium-bromide-stained gel before blotting is shown in the lower panel. (B) UPF1 protein from an aliquot of the same cells used in A was detected by Western blotting. Cell lysate corresponding to an equal number of cells was loaded in both lanes, and methylene blue staining of the membrane after blotting showed equal amounts of total protein in both lanes (not shown).
Computational predictions of the effect of PTC mutations on exonic splicing enhancers
Next, we decided to test more carefully whether alt-3′ ss usage is specifically induced when the reading frame is disrupted by a PTC, or whether alternatively the nonsense mutations disrupt exonic splicing enhancers (ESEs). Recently, two different computational methods have been developed that predict putative ESEs in a given RNA sequence: ESEfinder (version 2.0, http://rulai.cshl.edu/tools/ESE/; Cartegni et al. 2003) and RESCUE-ESE (http://genes.mit.edu/chris/; Fairbrother et al. 2002, 2004). We used both programs to scan the VDJ exon of miniμ and examined whether the nonsense mutations Ter32, Ter57, and Ter73 were predicted to alter putative ESEs. The A-to-T mutations at amino acid positions 32 and 57, which create Ter32 and Ter57, are in regions where for the wild-type sequence no binding sites for any of the four SR proteins were predicted by ESEfinder. In both cases, the mutation increases the score of a predicted binding site for SRp55 from 2.57 (just below threshold) to 3.16. In contrast, RESCUE-ESE predicts two overlapping ESEs spanning codon 32, both of which are eliminated by the A-to-T mutation. Similarly, six overlapping ESEs are predicted by RESCUE-ESE to span codon 57, and mutation of the A to T (nonsense) or G (missense) or C (missense) all are predicted to eliminate four of the six ESE hexamers. At codon 73, the G-to-T nonsense mutation decreases the score of an ESEfinder-predicted binding site for SC35 significantly from 2.51 (just above threshold) to 0.25, and RESCUE-ESE also predicts the elimination of an ESE by this mutation.
In summary, ESEfinder and RESCUE-ESE differ in their predictions about the effect of the nonsense mutations at codons 32 and 57 on ESEs, preventing us from drawing any conclusions. On the other hand, both programs predict that the nonsense mutation at codon 73 interferes with an ESE, suggesting that induction of alt-3′ splice site usage in Ter73 might not be frame dependent.
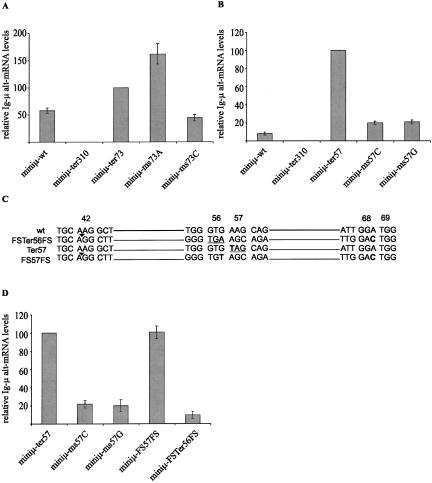
Alternative splicing of PTC+ Ig-μ transcripts is not frame dependent
To experimentally test whether disruption of the reading frame or disruption of ESEs is responsible for the alternative splicing of PTC+ miniμ transcripts, we made additional constructs with missense mutations at amino acid positions 57 or 73 (ms57C, ms57G, ms73A, and ms73C). When transiently expressed in HeLa cells, both missense mutations at amino acid position 73 induced NAS to an extent similar to that of the nonsense mutation (Fig. 4A), demonstrating that all changes in the first nucleotide of codon 73 induce NAS, irrespective of whether they truncate the reading frame or not. Consistent with the experimental result, ESEfinder predicts that not only mutation of the G to a T (Ter73), but also to an A or a C inactivates the putative SC35 binding site: The scores for ms73A and ms73C drop below the threshold to 2.06 and 0.25, respectively. Likewise, RESCUE-ESE predicts that not only the nonsense, but also the two missense mutations eliminate the putative ESE. Taken together, this suggests that changing the first nucleotide of codon 73 destroys a SC35 binding site, which leads to the inactivation of an ESE and to the activation of the alt-3′ splice site.
FIGURE 4.
Activation of the alt-3′ splice site does not correlate with premature termination of the reading frame. Total cellular RNA from HeLa cells transiently transfected with the indicated miniμ constructs were analyzed 48 h post-transfection by real-time RT-PCR as in Figure 2. The relative alt-mRNA level of Ter73 was set as 100% in A and of Ter57 in B and D. Missense mutations are named “ms” followed by the amino acid position and the changed nucleotide, for example, construct ms73A has the G at the first position of codon 73 changed to an A. (C) In constructs FSTer56FS and FS57FS, the reading frame between amino acid positions 42 and 68 was shifted by deletion of one nucleotide at position 42 (black triangle) and insertion of a C at position 68 (bold). These two frame-shift mutations eliminate the PTC in Ter57 (giving FS57FS) and create a PTC at amino acid position 56 in the wild-type construct (giving FSTer56FS). Except for miniμ-ter310, which served as a negative control, constructs with a −1 frame-shift (−1FS; see Fig. 1A) were used.
At amino acid position 57, the missense mutations ms57C and ms57G resulted in about fivefold less NAS than the Ter57, which at first glance is suggestive of frame dependence (Fig. 4B). However, we also noted that compared to wild-type transcripts, the missense mutations showed approximately twofold more alt-mRNA. ESEfinder predicts that the A-to-C mutation in ms57C generates a new SRp55 binding site (score 4.71) and a new SF2/ASF binding site (score 2.83), while the A-to-G mutation in ms57G causes no significant changes in the scores of any of the four SR proteins. As mentioned above, RESCUE-ESE in contrast predicts that all three point mutations destroy four of the six predicted ESE hexamer motifs that overlap with codon 57. Since for amino acid position 57, it was not possible to unambiguously distinguish between frame-dependent NAS and ESE-dependent NAS based on the data from the nonsense and the missense mutations, we introduced in Ter57 a +1 frame shift at amino acid position 42 and a −1 frame shift at amino acid position 68/69, which eliminates the PTC at amino acid position 57 (giving FS57FS). The same mutations in the wild-type construct create a PTC at amino acid position 56 (construct FSter56FS; Fig. 4C). As shown in Figure 4D, the PTC+ construct FSter56FS failed to induce NAS above the level detected with the wt construct, and the PTC- construct FS57FS still gave alt-mRNA levels similar to that of Ter57. This result unambiguously demonstrates that the induction of alt-mRNA is not frame dependent, but most probably results from a direct interference of the A-to-T mutation at amino acid position 57 with splicing. We conclude that NAS of miniμ induced by nonsense mutations at codons 57 and 73 is not PTC specific.
Conclusions
In summary, we found no evidence for reading-frame-dependent up-regulation of Ig-μ alt-mRNA at the two positions examined. Computational prediction of alterations of ESEs by the point mutations in miniμ using ESEfinder and RESCUE-ESE gave not entirely consistent results. The scores for specific SR protein binding sites predicted by the program ESEfinder did not decrease for several mutations that induced alt-3′ss usage. The predictions by RESCUE-ESE correlated better with our experimental results: With the exception of the G-to-C mutation at codon 73, all other mutations predicted to destroy one or more hexameric ESE motifs resulted in elevated alt-mRNA levels. Altogether, it seems most likely that the introduced mutations directly interfere with splicing signals required for proper exon definition. The finding that Ig-μ NAS occurs independently of NMD (see above) is also consistent with activation of the alt-3′ splice site being a direct consequence of ESE disruption.
Although we cannot exclude that PTCs at other, not tested positions would activate NAS in a frame-dependent manner, it seems unlikely that by coincidence we would have selected two positions for our analysis where disruption of splicing enhancers by the mutations would mask frame-dependent NAS. The results presented here show that frame-dependent NAS, if it exists at all, does not represent a general response to PTCs of transcripts encoded by genes of the immunoglobulin superfamily. These data together with our results from investigating NAS of TCR-β pre-mRNA, which in contrast to earlier reports do not reveal any evidence for frame dependence (Mohn et al. 2005), seriously question the idea that premature termination of the ORF can affect pre-mRNA splicing. Thus, in the absence of new, compelling evidence for frame-dependent NAS, we are reluctant to believe it exists.
MATERIALS AND METHODS
Cell lines and plasmids
The mouse Ig-μ hybridoma cell lines were previously described (Connor et al. 1994). The Ig-μ minigene and polyclonal HeLa cell populations stably expressing Ig-μ minigene constructs are described elsewhere (Bühler et al. 2004). Missense and frame-shift mutations in the Ig-μ minigene were generated by PCR-mediated site-directed mutagenesis using the QuikChange XL Site-Directed Mutagenesis Kit (Stratagene). Oligonucleotide sequences used are available on request. All mutations were confirmed by sequencing the relevant parts of the constructs. Mutation of codon 73 (GGA) to AGA and CGA resulted in constructs ms73A and ms73C, and mutation of codon 57 (AAG) to GAG and CAG gave constructs ms57G and ms57C, respectively. Constructs denoted as −1FS were obtained by insertion of an adenosine between codons 104 and 105, generating a −1 frame shift. Constructs FSTer56FS and FS57FS were derived from Ig-μ wt and Ter57, respectively, by deletion of the first adenosine in codon 42 (+1 frame shift) and insertion of a cytosine between codons 68 and 69 (−1 frame shift; see Fig. 4C). Conditions for cell culture and transfections were as previously described (Bühler et al. 2004).
Real-time RT-PCR
Conditions for reverse transcription of total cellular RNA and for real-time PCR were as previously described (Bühler et al. 2004). Ig-μ alt-mRNA was measured over the junction between the 5′ splice site of the leader exon and the alt-3′ splice site using 400 nM 5′-GATGGAGCCGGATCTTTCTCT-3′, 400 nM 5′-AGGCTGC TGAGCTGCATGT-3′ and 200 nM 5′-FAM-CCTCCTGTCAATA ATTGCAGCACAGCC-TAMRA-3′. Primer and TaqMan probe used to measure neomycin mRNA was described previously (Bühler et al. 2004), and 18S rRNA was measured using predeveloped assay reagents from Applied Biosystems.
RT-PCR and cloning of alt-mRNA
Total RNA from cells expressing either miniμ-ter73 or -ter310 was isolated and reverse transcribed as for real-time RT-PCR, except that 6 ng/μL of oligo 5′-(dT)30VN-3′ were used instead of random hexamers. Reverse transcribed material corresponding to 100 ng RNA was amplified with the PCR Core Kit (Qiagen) in 50 μL PCR buffer containing 200 nM dNTPs, 5 U Taq polymerase, and 244 nM forward (5′-ACTGCATTGTCGACCTCACCATGG GATGGAG-3′) and reverse primer (5′-GGGATGGTGAAGGTTA GGATGTC-3′), which are complementary to the leader exon and the constant region exon C3, respectively. The RT-PCR reaction was separated on a 2% agrose gel and the alt-mRNA band specific for miniμ-ter73 was gel purified and cloned into the TOPO vector pCRII (Invitrogen) for subsequent sequencing.
For the RT-PCR shown in Figure 1C, the same forward primer as above was used together with a reverse primer complementary to the constant region exon C1 (5′-AGCGCCTAGGGAGGTGGC TAGGTACTTGCCCCCTG-3′).
Northern blot analysis
Northern blot analysis was performed as previously described (Bühler et al. 2004) using [α-32P]-labeled miniμ-ter310 DNA as a probe.
Western blot analysis
Whole-cell lysate corresponding to 105 cells per lane was electrophoresed on a 10% SDS PAGE. Protein was transferred to Optitran BA-S 85 reinforced nitrocellulose (Schleicher & Schuell) and probed with a 1:500 dilution of affinity purified goat anti-mouse IgM (μ chain-specific, Jackson ImmunoResearch Laboratories) to detect IgM polypeptide. The primary antibody was detected with a 1:4000 diluted horseradish peroxidase (HRP)-conjugated donkey anti-goat IgG antibody (Promega) using the ECL+ Western blotting detection system (Amersham Pharmacia Biotech). hUpf1 was detected with a 1:2500 diluted polyclonal rabbit antibody (gift from Jens Lykke-Andersen, Univ. of Colorado, Boulder) and Lamin C was detected with a 1:1000 diluted monoclonal mouse antibody (Santa Cruz Biotechnology). Secondary antibodies were 1:2500 diluted HRP conjugated α-rabbit or α-mouse, respectively.
RNAi
Knock down of hUpf1 by RNAi was induced by cotransfection of pEGFP-N1 (Invitrogen) and a pSUPER vector (Brummelkamp et al. 2002) targeting the sequence GAGAATCGCCTACTTCACT using Lipofectamine (Invitrogen) as described above. Seventy-two hours post-transfection, GFP-expressing cells were collected by flow cytometry, and protein and total RNA were isolated. Western and Northern blot analyses were performed as described above.
Acknowledgments
We are grateful to Jens Lykke-Andersen for anti-hUpf1 antibody. This work was financed by the Swiss National Research Foundation (grants 3100-61720.00 and 3100A0-102159), the Kanton Bern, the Novartis Foundation for Biomedical Research, and the Helmut Horten Foundation.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7183805.
REFERENCES
- Aoufouchi, S., Yelamos, J., and Milstein, C. 1996. Nonsense mutations inhibit RNA splicing in a cell-free system: Recognition of mutant codon is independent of protein synthesis. Cell 85: 415–422. [DOI] [PubMed] [Google Scholar]
- Baumann, B., Potash, M.J., and Kohler, G. 1985. Consequences of frameshift mutations at the immunoglobulin heavy chain locus of the mouse. EMBO J. 4: 351–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe, B.J. 2000. Exonic splicing enhancers: Mechanism of action, diversity and role in human genetic diseases. Trends Biochem. Sci. 25: 106–110. [DOI] [PubMed] [Google Scholar]
- Brummelkamp, T.R., Bernards, R., and Agami, R. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296: 550–553. [DOI] [PubMed] [Google Scholar]
- Buchner, D.A., Trudeau, M., and Meisler, M.H. 2003. SCNM1, a putative RNA splicing factor that modifies disease severity in mice. Science 301: 967–969. [DOI] [PubMed] [Google Scholar]
- Bühler, M., Paillusson, A., and Mühlemann, O. 2004. Efficient down-regulation of immunoglobulin μ mRNA with premature translation-termination codons requires the 5′-half of the VDJ exon. Nucleic Acids Res. 32: 3304–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputi, M., Kendzior Jr., R.J., and Beemon, K.L. 2002. A nonsense mutation in the fibrillin-1 gene of a Marfan syndrome patient induces NMD and disrupts an exonic splicing enhancer. Genes & Dev. 16: 1754–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni, L., Chew, S.L., and Krainer, A.R. 2002. Listening to silence and understanding nonsense: Exonic mutations that affect splicing. Nat. Rev. Genet. 3: 285–298. [DOI] [PubMed] [Google Scholar]
- Cartegni, L., Wang, J., Zhu, Z., Zhang, M.Q., and Krainer, A.R. 2003. ESEfinder: A Web resource to identify exonic splicing enhancers. Nucleic Acids Res. 31: 3568–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter, M.S., Doskow, J., Morris, P., Li, S., Nhim, R.P., Sandstedt, S., and Wilkinson, M.F. 1995. A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J. Biol. Chem. 270: 28995–29003. [DOI] [PubMed] [Google Scholar]
- Carter, M.S., Li, S., and Wilkinson, M.F. 1996. A splicing-dependent regulatory mechanism that detects translation signals. EMBO J. 15: 5965–5975. [PMC free article] [PubMed] [Google Scholar]
- Connor, A., Wiersma, E., and Shulman, M.J. 1994. On the linkage between RNA processing and RNA translatability. J. Biol. Chem. 269: 25178–25184. [PubMed] [Google Scholar]
- Dahlberg, J.E., Lund, E., and Goodwin, E.B. 2003. Nuclear translation: What is the evidence? RNA 9: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz, H.C., Kendzior Jr., R.J. 1994. Maintenance of an open reading frame as an additional level of scrutiny during splice site selection. Nat. Genet. 8: 183–188. [DOI] [PubMed] [Google Scholar]
- Fairbrother, W.G., Yeh, R.F., Sharp, P.A., and Burge, C.B. 2002. Predictive identification of exonic splicing enhancers in human genes. Science 297: 1007–1013. [DOI] [PubMed] [Google Scholar]
- Fairbrother, W.G., Yeo, G.W., Yeh, R., Goldstein, P., Mawson, M., Sharp, P.A., and Burge, C.B. 2004. RESCUE-ESE identifies candidate exonic splicing enhancers in vertebrate exons. Nucleic Acids Res. 32: W187–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gersappe, A. and Pintel, D.J. 1999. A premature termination codon interferes with the nuclear function of an exon splicing enhancer in an open reading frame-dependent manner. Mol. Cell. Biol. 19: 1640–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley, B.R. 2000. Sorting out the complexity of SR protein functions. RNA 6: 1197–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudikote, J.P. and Wilkinson, M.F. 2002. T-cell receptor sequences that elicit strong down-regulation of premature termination codon-bearing transcripts. EMBO J. 21: 125–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze, M.W. and Kulozik, A.E. 1999. A perfect message: RNA surveillance and nonsense-mediated decay. Cell 96: 307–310. [DOI] [PubMed] [Google Scholar]
- Li, S. and Wilkinson, M.F. 1998. Nonsense surveillance in lymphocytes? Immunity 8: 135–141. [DOI] [PubMed] [Google Scholar]
- Li, B., Wachtel, C., Miriami, E., Yahalom, G., Friedlander, G., Sharon, G., Sperling, R., and Sperling, J. 2002. Stop codons affect 5′ splice site selection by surveillance of splicing. Proc. Natl. Acad. Sci. 99: 5277–5282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H.X., Cartegni, L., Zhang, M.Q., and Krainer, A.R. 2001. A mechanism for exon skipping caused by nonsense or missense mutations in BRCA1 and other genes. Nat. Genet. 27: 55–58. [DOI] [PubMed] [Google Scholar]
- Lozano, F., Maertzdorf, B., Pannell, R., and Milstein, C. 1994. Low cytoplasmic mRNA levels of immunoglobulin κ light chain genes containing nonsense codons correlate with inefficient splicing. EMBO J. 13: 4617–4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat, L.E. 2001. The power of point mutations. Nat. Genet. 27: 5–6. [DOI] [PubMed] [Google Scholar]
- ———. 2004. Nonsense-mediated mRNA decay: Splicing, translation and mRNP dynamics. Nat. Rev. Mol. Cell. Biol. 5: 89–99. [DOI] [PubMed] [Google Scholar]
- Mendell, J.T., ap Rhys, C.M., and Dietz, H.C. 2002. Separable roles for rent1/hUpf1 in altered splicing and decay of nonsense transcripts. Science 298: 419–422. [DOI] [PubMed] [Google Scholar]
- Mohn, F., Bühler, M., and Mühlemann, O. 2005. Nonsense-associated alternative splicing of T-cell receptor β genes: No evidence for frame dependence. RNA (this issue). [DOI] [PMC free article] [PubMed]
- Mühlemann, O., Mock-Casagrande, C.S., Wang, J., Li, S., Custódio, N., Carmo-Fonseca, M., Wilkinson, M.F., and Moore, M.J. 2001. Precursor RNAs harboring nonsense codons accumulate near the site of transcription. Mol. Cell 8: 33–43. [DOI] [PubMed] [Google Scholar]
- Pagani, F., Buratti, E., Stuani, C., and Baralle, F.E. 2003. Missense, nonsense, and neutral mutations define juxtaposed regulatory elements of splicing in cystic fibrosis transmembrane regulator exon 9. J. Biol. Chem. 278: 26580–26588. [DOI] [PubMed] [Google Scholar]
- Valentine, C.R. 1998. The association of nonsense codons with exon skipping. Mutat. Res. 411: 87–117. [DOI] [PubMed] [Google Scholar]
- Wang, J., Chang, Y.F., Hamilton, J.I., and Wilkinson, M.F. 2002a. Nonsense-associated altered splicing: A frame-dependent response distinct from nonsense-mediated decay. Mol. Cell 10: 951–957. [DOI] [PubMed] [Google Scholar]
- Wang, J., Gudikote, J.P., Olivas, O.R., and Wilkinson, M.F. 2002b. Boundary-independent polar nonsense-mediated decay. EMBO Rep. 15: 274–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Hamilton, J.I., Carter, M.S., Li, S., and Wilkinson, M.F. 2002c. Alternatively spliced TCR mRNA induced by disruption of reading frame. Science 297: 108–110. [DOI] [PubMed] [Google Scholar]
- Wilkinson, M.F. and Shyu, A.B. 2002. RNA surveillance by nuclear scanning? Nat. Cell. Biol. 4: E144–E147. [DOI] [PubMed] [Google Scholar]