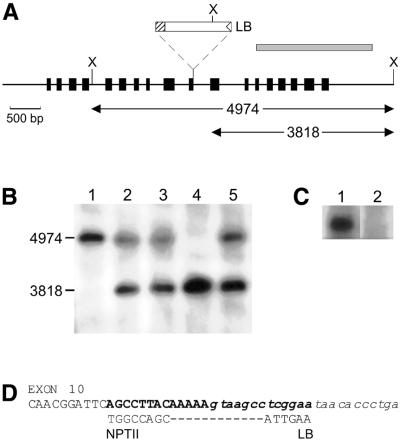
Figure 2.
(A) Genomic organisation of the AtKU70 locus with the insertion point of the T-DNA indicated. The exons of AtKU70 are shown as boxes. The probe used for the DNA blots was a 1871 bp PCR fragment amplified using the primers AK1 + AK3 and is represented as a shaded box. X, XhoI restriction sites. LB, T-DNA left border. The truncated NPTII ORF on the T-DNA is shown as a hatched box. (B) DNA blot. Lane 1, wild-type (Ws) seedlings; lanes 2–5, individual plants containing a T-DNA in AtKU70. (C) Northern analysis: 8 µg total RNA from 12-day-old seedlings was blotted and probed using the 1.8 kb AtKU70 cDNA. Lane 1, Ws; lane 2, AtKU70–/– seedlings. (D) Integration site of the T-DNA. Upper line, the sequence of exon 10 of the AtKU70 gene is shown in uppercase letters. Intron 10 is indicated in lowercase italics. The sequence deleted due to the T-DNA insertion is in bold. Middle line, the ends of the T-DNA are shown. The left T-DNA border (LB) had lost 9 bp from its end and was integrated into intron 10. The T-DNA right border (RB) end was heavily truncated (∼4 kb lost) resulting in part of the NPTII ORF being fused to AtKU70 exon 10.