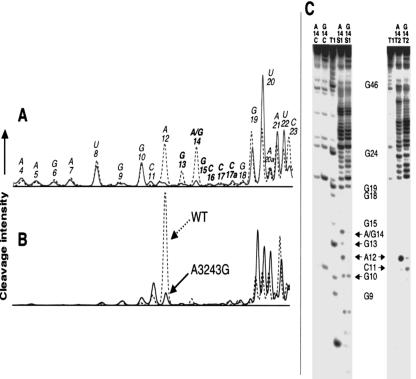
FIGURE 2.
Enzymatic probing of the wild-type (WT) and mutant A3243G hs mt tRNALeu(UUR). (A) Normalized histograms generated from PAGE analysis of enzymatic cleavage of wild-type (dashed) and mutant A3243G (solid) hs mt tRNALeu(UUR) using nuclease S1 under nondenaturing conditions. Positions are numbered according to the consensus tRNA numbering system, and bold nucleotides are located within the proposed dimer interface. (B) Histograms generated from PAGE analysis of enzymatic cleavage of wild-type (dashed) and mutant A3243G (solid) hs mt tRNALeu(UUR) using RNase T2. (C) 20% denaturing PAGE of nuclease S1 and RNase T2 structural probing experiments. Sample lanes marked A14 represent probing of the wild-type hs mt tRNALeu(UUR), while those labeled G14 represent probing results for the mutant A3243G (A14G) hs mt tRNALeu(UUR). Lanes marked T1 represent the guanine ladder performed on wild-type hs mt tRNALeu(UUR). Lanes marked C are control samples that were not incubated with nucleases. Positions where losses in cleavage efficiency are observed due to dimerization in the mutant structure are identified with arrows. Multiple trials (> 3) of all probing experiments were performed to ensure the reproducibility of the trends shown.