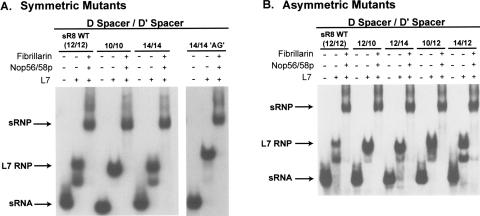
FIGURE 3.
Alteration of the spacing distance between the sR8 box C/D and C′/D′ motifs does not affect sRNP assembly. sR8 sRNA D and D′ spacer regions were shortened or lengthened in 2-nt increments by deleting or inserting nucleotides upstream and downstream of boxes C′ and C, respectively, as indicated in Figure 1 ▶. Spacing mutants lengthening the D and D′ spacers were typically created by inserting two uridine nucleotides unless otherwise indicated (spacing mutant 14/14 AG inserted the dinucleotide AG in place of UU). sR8 spacing mutants were either symmetric (A), where both the D and D′ spacers were shortened/lengthened, or asymmetric (B), where only one spacer region was altered. Altered spacing distances are indicated at the top (D spacer/D′ spacer). sR8 sRNPs were assembled in vitro and analyzed by electrophoretic mobility-shift analysis. The sequential addition of the box C/D sRNP core proteins to radiolabeled wild-type and mutant spacer sR8 RNAs is indicated at the top and the migration positions of the resulting RNP complexes are designated at the side. The slightly altered migration of the 14/14 (AG) RNPs is a result of slightly different electrophoretic conditions.