FIGURE 2.
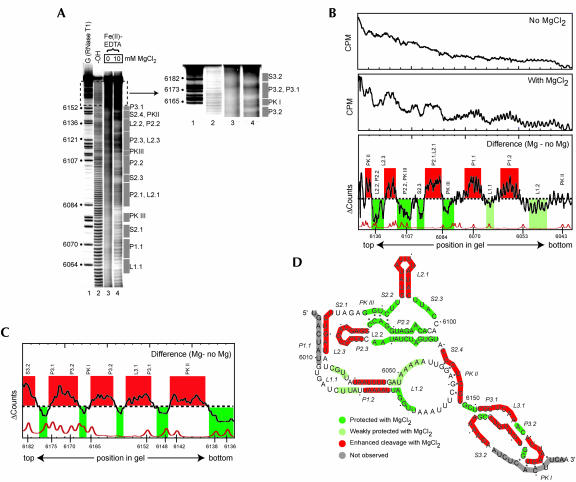
Hydroxyl radical probing of the IGR IRES. (A) Example of a hydroxyl radical probing experiment performed on 5′ end-labeled PSIV IRES RNA. Lanes 3 and 4 contain the probed RNA in the absence and presence of MgCl2. The top portion of the gel is expanded and the contrast adjusted to show the cleavage pattern near the 3′ end of the RNA. The location of various secondary structural domains are noted to the right of the gel. Lane 1 contains an RNase T1 ladder that is annotated on the left and lane 2 is a partial hydrolysis ladder. (B) Quantitative analysis of the hydroxyl radical probing profile of a portion of PSIV IRES RNA. Gel lanes were quantitated as a function of position in the gel, normalized, and compared as described in the text. The top is a lane with no added magnesium, the center is a lane with 10 mM added magnesium, and the bottom is the calculated difference. A trace of the RNase T1 sequencing lane (bottom) allows precise identification of the locations of the protections on the RNA backbone. Regions that we assigned as protected upon folding are shown in green, with weaker protections in light green. Regions that show enhanced cleavage are shown with red. The locations of corresponding secondary structures features are shown above the trace. (C) Results of an analysis identical to part B on a gel that resolved the portion of the RNA backbone corresponding to region 3 at the 3′ end of the PSIV IRES RNA. (D) Map of hydroxyl radical cleavage profile on the PSIV IRES secondary structure. Color scheme is identical to that in B and C. Gray areas denote parts of the backbone not observed on the gel.