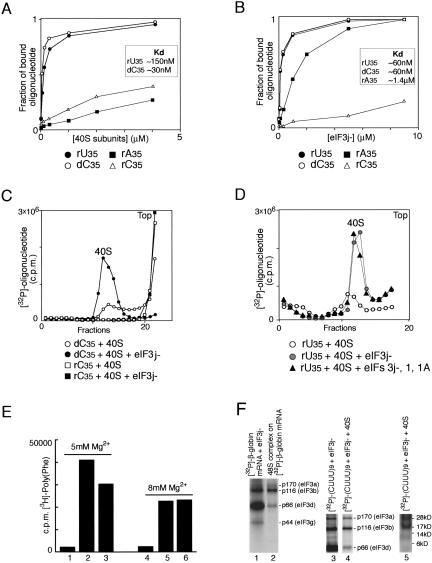
FIGURE 3.
Binding of oligonucleotides to eIF3j− and to 40S subunits. (A,B) Affinity measurements made in filter-binding experiments using (A) 40S subunits and (B) eIF3j− with [32P]-labeled oligonucleotides as indicated. The fraction of bound oligonucleotide is the ratio of [32P]-labeled oligonucleotide retained on the filter to the input. Dissociation constants are shown in the inset boxes. (C) Binding of [32P]-labeled oligonucleotide (dC35 or rC35) to 40S subunits in the presence and absence of eIF3j−. (D) Influence of eIF1 and eIF1A on binding of [32P]-labeled rU35 to the 40S subunit in the presence of eIF3j−. Ribosomal complexes were separated by centrifugation in 10%–30% linear sucrose gradients. Sedimentation was from right to left. The position of 40S subunits determined by optical density is indicated. Upper fractions from the gradient have been omitted for clarity. (E) Template-dependent [3H]-polyphenylalanine synthesis by ribosomes assembled by incubation at 5 mM Mg2+ (lanes 1–3) or 8 mM Mg2+ (lanes 4–6) of 40S and 60S subunits without poly(U) (lanes 1,4), 60S subunits with preassembled 40S subunit/poly(U) complexes (lanes 2,5) or poly(U) with preassembled 80S ribosomes (lanes 3,6). All reaction mixtures contained [3H]-Phe-tRNAPhe, EF1A, and EF2. [3H]-polyphenylalanine was counted after TCA precipitation of reaction mixtures on nitro-cellulose filters. (F) UV-crosslinking of [32P]-β-globin mRNA (lanes 1,2) and [32P]-(CUUU)9 (lanes 3–5) to eIF3j− (lanes 1–4) and 40S subunits (lane 5) in binary eIF3j−/RNA complexes (lanes 1,3), 48S complexes (lane 2), and eIF3j−/(CUUU)9/40S subunit complexes (lanes 4,5). Polypeptides resolved by gel electrophoresis were visualized by autoradiography. eIF3a (p170), eIF3b (p116), eIF3d (p66), and eIF3g (p44) are indicated to the right of lanes 2,4. The positions of molecular weight markers are shown to the right of the radiolabeled ribosomal proteins in lane 5.