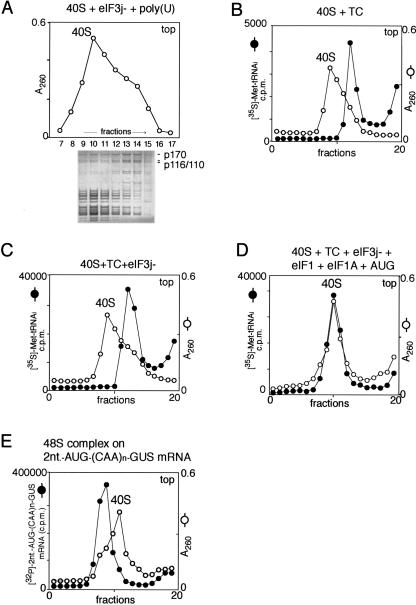
FIGURE 6.
Mobility of ribosomal complexes during centrifugation through 10%–30% linear sucrose density gradients. (A) Fractionation by sucrose density gradient centrifugation of eIF3j−/poly(U)/40S subunit complexes formed with excess 40S subunits. The positions of 40S subunits and their complexes with eIF3j− were determined by optical density. Polypeptides in different fractions were resolved by electrophoresis followed by Coomassie staining. eIF3a (p170) and eIF3b/3c (p116/p110) are labeled to the right of the lower panel. (B,C) Mobility of preinitiation 43S complexes (consisting of 40S subunits and eIF2-ternary complex [TC]) with (C) or without (B) eIF3, compared to free 40S subunits. (D) Mobility of 43S preinitiation complexes (consisting of 40S subunits, eIF1, eIF1A, TC, eIF3, and AUG triplets) compared to free 40S subunits. The positions of 40S subunits and their complexes with TC, eIF3, eIF1, and eIF1A were determined by optical density and by scintillation counting of [35S]Met-tRNAiMet (B,C,D). (E) Mobility of 48S initiation complexes assembled on [32P]-2-nt-AUG-(CAA)n-GUS mRNA compared to free 40S subunits. The positions of 40S subunits and 48S complexes were determined by optical density and by Cherenkov counting of [32P]-2-nt-AUG-(CAA)n-GUS mRNA. Sedimentation was from right to left. Upper fractions from the gradient have been omitted for clarity.