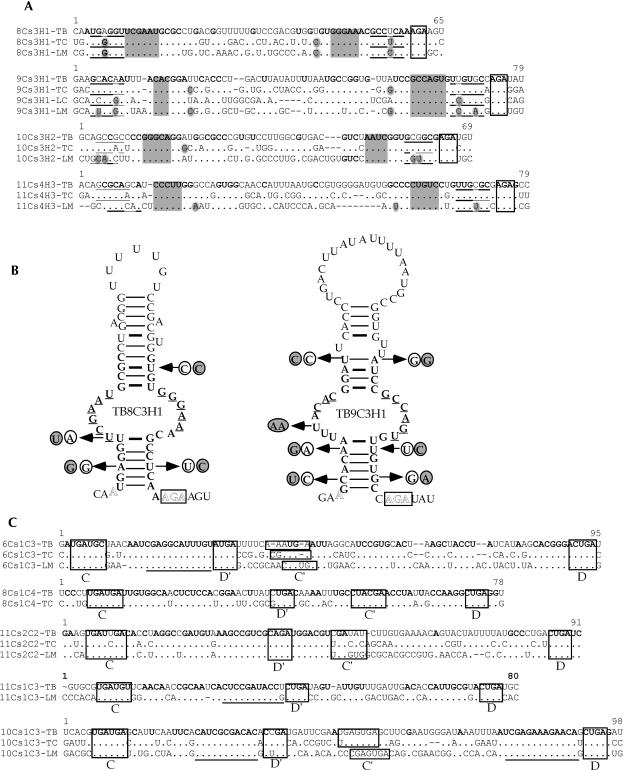
FIGURE 4.
Sequence alignment of snoRNAs from different trypanosomatid species. (A) Examples of H/ACA RNAs. The AGA motif is boxed. The gaps are shown as short lines, whereas conserved residues are given in dots. The nucleotides conserved in all species are in bold. The species designations are (TB) Trypanosoma brucei, (TC) Trypanosoma cruzi, and (LM) Leishmania major. Domains involved in base-pairing with their targets are shadowed. (B) Secondary structure of H/ACA-like RNA, TB8Cs3H1, and TB9Cs3H1. The sequences of the RNA are from T. brucei. Compensatory changes between these sequences among the trypanosomatid species are marked by arrows. The sequences of T. cruzi are circled, and the L. major sequences are circled and shadowed. (C) Examples of C/D snoRNAs. The C/D- and C′/D′ boxes are indicated. The sequences involved in base-pairing with target RNAs are underlined.