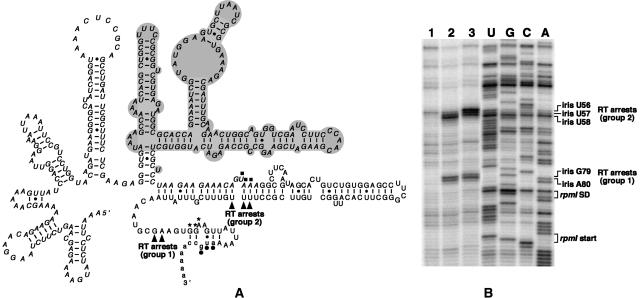
FIGURE 6.
L20 and L20ΔN-induced RT arrests are located at similar positions on rpmI mRNA. (A) Secondary structure of the rpmI translational operator, i.e., the region upstream of rpmI mRNA containing the two L20-binding sites required for control (Chiaruttini et al. 1996). Nucleotide residues forming the infC stop codon and the rpmI Shine–Dalgarno (SD) sequence and start codon are indicated by filled squares, asterisks, and dots, respectively. The nucleotide sequences of infC, iris (iris is an acronym for the infC-rpmI intergenic sequence) and rpmI are in italics, uppercase and lowercase, respectively. The ΔApaLI deletion which does not affect control (Chiaruttini et al. 1996) is in gray background. Arrowheads indicate the two groups (groups 1 and 2) of RT arrests caused by L20 and L20ΔN. (B) Primer-extension assay. A shortened form of the operator in which nucleotide residues corresponding to the ΔApaLI deletion have been removed was used in this experiment. (Lane 1) rpmI operator alone; (lane 2) rpmI operator with a 30-fold molar excess of wild-type L20; (lane 3) rpmI operator with a 90-fold molar excess of L20ΔN. The positions of the L20- and L20ΔN-induced RT arrests and those of the nucleotide residues forming the rpmI SD sequence and start codon are indicated to the right of the autoradiogram.