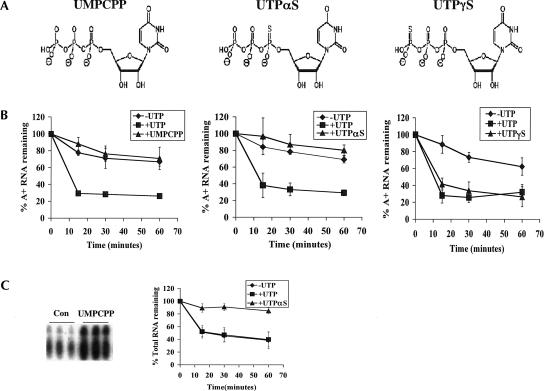
FIGURE 4.
The effect of nonhydrolyzable analogs on degradation of polyadenylated RNAs. (A) Structures of the nonhydrolyzable analogs used in this assay. (From left to right) UMPCPP [uridine-5-O-(α,β-methylene-triphosphate)], UTPαS [uridine-5′-(α-thio)-triphosphate], and UTPγS [uridine-5′-(γ-thio)-triphosphate]. (B) Mitochondria were labeled with [α-32P]CTP and subsequently incubated in transcription buffer containing 2 mM CTP (-UTP, diamonds), 2 mM CTP and 2 mM UTP (+UTP, squares), or 2 mM CTP and 2 mM UTP analog [triangles; +UMPCPP (left), +UTPαS (middle), and +UTPγS (right)]. At indicated time points, incorporation of [α-32P]CTP into polyadenylated RNAs was measured. Percent polyadenylated RNA remaining as compared to pulse-labeled RNA is plotted. Each point is the average of three experiments with error bars representing one standard deviation. (C, left panel) Mitochondria were pulsed in triplicate with [α-32P]CTP in the presence of 0.1 mM GTP, UTP, and ATP (Con) or these nucleotides and 2 mM UMPCPP. After purification, total RNA was resolved on a 7 M urea and 6% acrylamide gel and visualized by autoradiography. The region of the gel at which tRNAs migrate is shown. (C, right panel) Mitochondria were labeled with [α-32P]CTP and subsequently incubated in transcription buffer containing 2 mM CTP (-UTP, diamonds), 2 mM CTP and 2 mM UTP (+UTP, squares), or 2 mM CTP and 2 mM UTPαS (+UTPαS, triangles). At indicated times, incorporation of [α-32P]CTP into total RNAs was measured. Percent total RNA remaining as compared to pulse-labeled RNA is plotted.