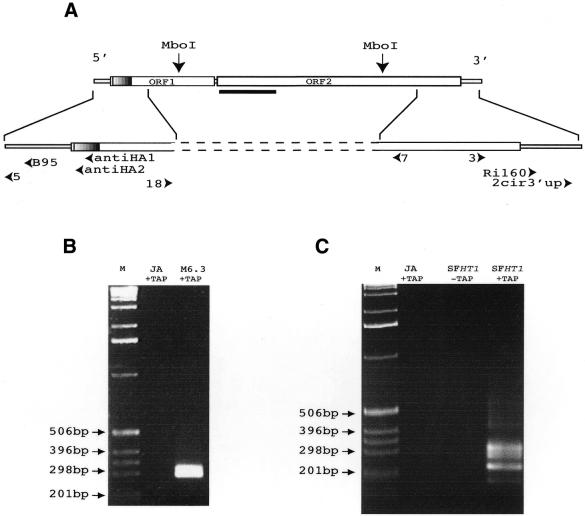
Figure 1.
Analyses of the transcripts produced by the I-HA-O1-Δnuc and I-HA-O1 elements. (A) Structure of the I-HA-O1 and I-HA-O1Δnuc elements. The two large open boxes represent the two ORFs of the I-HA-O1 element, and the small open boxes represent the 5′ and 3′ UTRs. The small hatched region at the beginning of ORF1 indicates the position of the HA epitope which is inserted in frame with the rest of ORF1 (11). The most 5′ and 3′ MboI restriction sites that were used for inverse PCR are indicated. The bold line below the beginning of ORF2 indicates the position of the deletion in I-HA-O1-Δnuc. Arrowheads indicate the positions and orientations of the primers used in RT–PCR and sequencing experiments (see Materials and Methods). (B and C) Analyses on a 1.5% agarose gel of the final products of the RNA circularization/RT–PCR experiments. (B) RNAs were extracted from ovaries of JA females and M6.3 females, transgenic for I-HA-O1-Δnuc. (C) RNAs were extracted from ovaries of JA females and SFHT1 females, issued from crosses between JA females and inducer males of the HT1 line containing five I-HA-O1 elements. RNAs were treated (+TAP) or not (–TAP) with TAP prior to intra-molecular ligation. Reverse transcription was primed using primer antiHA1, and the resulting cDNAs were submitted to two successive PCRs: the first, using primer pair antiHA2/RI160, and the second, using primer pair B95/2cir3′up. M, molecular markers (1 kb ladder from GibcoBRL™).