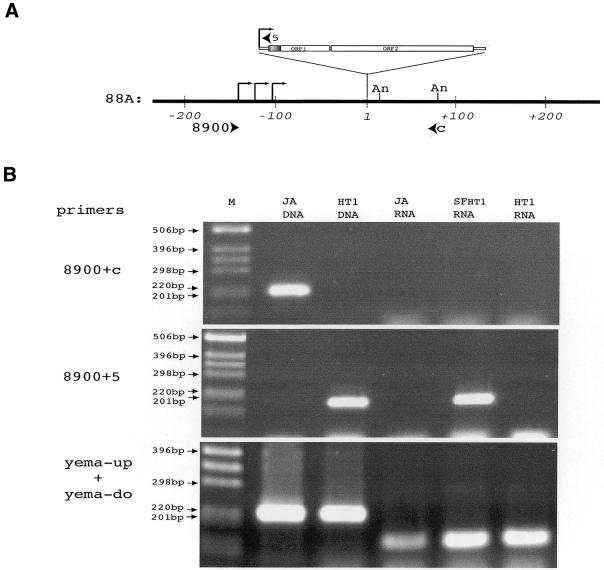
Figure 3.
Transcription of the I-HA-O1(88A) element. (A) Map of the 88A locus around the I-HA-O1(88A) insertion site. Coordinates on genomic DNA are indicated in base pairs, the first nucleotide of the target site duplication generated by the I-HA-O1 insertion being arbitrarily chosen as the origin. The I-HA-O1 element insertion is not shown to scale. The positions of transcription start sites of the transcripts of I-HA-O1(88A) shown in Figure 2B are indicated with broken arrows, and the positions of the polyadenylation sites are indicated with An. Arrowheads indicate the positions and orientations of the primer pairs 8900/5 and 8900/C used for RT–PCR. (B) RT–PCR analysis of the transcripts of the 88A region. RT–PCRs were performed on RNAs extracted from ovaries of JA, SFHT1 and HT1 females as described in Materials and Methods, using random hexamers to prime reverse transcription and primer pairs 8900/C (specific for the empty 88A locus), 8900/5 (specific for the 88A locus containing the I-HA-O1 insertion) and yema-up/yema-do (specific for the gene encoding yemanuclein-α) for PCR amplifications. Genomic DNA extracted from JA and HT1 adult flies were used for PCR controls. PCR products were analyzed on 1.5% agarose gels. M, molecular markers (1 kb ladder from GibcoBRL™).