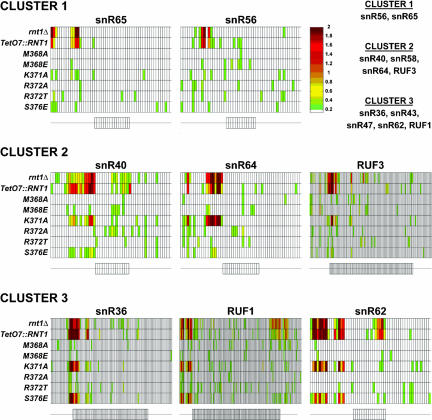
FIGURE 3.
Amino acid substitutions in Rnt1p dsRBD induce substrate-specific processing defects in vivo. Microarray analysis of the accumulation of known substrates of Rnt1p in strains lacking Rnt1p (rnt1Δ and TetO7::RNT1) and strains expressing the M368A, M368E, K371A, R372A, R372T, and S376E dsRBD mutant versions of Rnt1p. The small rectangles aligned in one row correspond to the relative fluorescence of RNA bound to oligonucleotide probes complementary to RNA sequences in the mature or flanking sequences of the snoRNA mentioned above. A schematic representation of the snoRNAs and their flanking sequences are shown below each set of data. On these schematics, rectangles correspond to probes complementary to the mature sequences of the snoRNAs, and thin lines correspond to probes complementary to the 5′ and 3′ flanking regions. The color of each rectangle is indicative of the relative accumulation of the corresponding RNA sequence in the mutant strain indicated on the left compared to the wild-type strain. The color scale is shown on the right; the reported values correspond to the log2 of the ratio over wild type of the fluorescent intensities. The snoRNA precursor substrates can be clustered into three main categories depending on their sensitivities to the Rnt1p dsRBD mutations. Cluster 1 corresponds to substrates that are insensitive to all tested mutations. Cluster 2 corresponds to substrates affected only by the K371A substitution, and Cluster 3, to substrates sensitive to both the K371A and S376E substitutions. The full set of substrates for each cluster is indicated on the upper right part of the figure.